Gene
KWMTBOMO15397
Annotation
PREDICTED:_uncharacterized_protein_LOC105841331_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.094
Sequence
CDS
ATGAAGACAGTTAAGCGTCTCGCCCGCGATGAAACTGATTTCCCGATCGCTTCAGAAATCGCTAGTGATCATCTTTATATGGATGATCTAGGAGTTTCGGTAGATACGCCCGAGAATGGAATTCAGTTGGCTCATGATTTAATCGAACTATTCAAACGTGGTGGTTTCGACTTAGTTAAATTCGTTAGCAATTCGCCCGAGGTGTTAAAATCGATTCCGCAAGCCAACCGCTTATCTTCAAAAATTGAATTCGACAAGGAGGATAATATGAAGCTCCTAGGTCTAATTTGGTCACCGGTCGAGGATGAATTTTCAATCGCTATTAAATTCAACCCGCAAGCAAAATGTACAAAGCGCCTTATGCTTTCCACAATCGCTAAACTGTGGGACATAAACGGATTTTGTGCCCCTGTCATCTTACACGCAAAATTATTAATAAAAGCTCTCTGGGTAGCTCAGATTGATTGGGATGCAGAGCCTCCAAGCGACATCTGTGCTGCGTGGAAGCGCTTTCTTAATGAACTCCCCGCTCTGCAAGACTTTAAAATACCTCGCTTCGTAGGCATCGTGCCCGAGTCCCGTGTCATACTACTGGGATTTGCGGACGCTTCTGAAAAAGCGATGGGTGCTTCTGTGTATATTTGTGTACAACCGGCTTCCGGACCAAATGAAGTCAATTTAATTGGTGCCAAAAGCAAGGTCGCGCCGCTCAAGGTTATTTCCTTAGCCCGCCTAGAGCTTTGTGCTGCTGTATTATTATCGAAACTGTTTCGATTTGTTATCGACACTGTTTCTCCCCGCTGTCATGTCGACGAAATTTTCGCGTTTACTGATTCGACAGTGACCCTCCATTGA
Protein
MKTVKRLARDETDFPIASEIASDHLYMDDLGVSVDTPENGIQLAHDLIELFKRGGFDLVKFVSNSPEVLKSIPQANRLSSKIEFDKEDNMKLLGLIWSPVEDEFSIAIKFNPQAKCTKRLMLSTIAKLWDINGFCAPVILHAKLLIKALWVAQIDWDAEPPSDICAAWKRFLNELPALQDFKIPRFVGIVPESRVILLGFADASEKAMGASVYICVQPASGPNEVNLIGAKSKVAPLKVISLARLELCAAVLLSKLFRFVIDTVSPRCHVDEIFAFTDSTVTLH
Summary
Uniprot
A0A1S3DJC4
S4NIN9
A0A1S3DBQ3
A0A1S3DTS8
A0A1Y1LIC8
A0A1S4EIX5
+ More
A0A1S3DMK8 A0A023F096 A0A0A9W266 A0A0A9VTB0 A0A1S3DIZ2 A0A0A9VUS3 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A1B6M7K2 A0A0A9X9A3 J9JUM9 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6LLT3 A0A1B6MT91 V5GC07 A0A1S3DMQ9 A0A1S3DM22 A0A3Q0JFQ3 A0A3Q0J0P0 V5GHM7 A0A182I0G9 A0A1S3DMT1 A0A1B6KIX6 A0A3Q0JHG9 A0A3Q0JJJ7 A0A146M0N4 A0A3Q0JBZ9 A0A3Q0J7K9 A0A3Q0J7N1 A0A1Y1LM33 A0A0A9XZV3 A0A1Y1LIG5 A0A0J7K8B6 D6WY83 W8BU98 A0A0J7K7U0 A0A2S2NBR8 A0A1B6GGX5 J9JNP4 A0A2S2QTX5 A0A182WES7 A0A146KZL4 X1X9U0 A0A3Q0IZW4 A0A0J7N391 A0A1S3DBK1 A0A0J7KGQ6 A0A1S3DQE7 W8AH72 A0A0J7N465 A0A0A9Z0W7 X1WPD9 A0A0A9WWW3 A0A2S2NNC8 A0A1B6MIG8 X1XET3 A0A0J7N5Z0 A0A0J7KH73 A0A224XGH3 A0A0A9WHD6 A0A1B6MUZ9 J9JUK4 A0A146KRI6 A0A0J7K952 A0A0J7K9I5 D7EKN2 A0A0J7KNK6 A0A1Y1LBZ3 X1WVF9 J9K0A1 A0A1B6KZV2 A0A2S2NDE3 A0A2S2QWC4 A0A2S2NHU0 A0A0N0PAG9 X1XU26 A0A0V0Z0L1 A0A0A9W8L8 X1X8C2 A0A2S2PT56 X1XTW5 A0A0A9VYL4 A0A0A9YD88 A0A0J7MYK0 A0A146LBU6 A0A0A9VSF7 A0A0J7KC70 X1XB32 A0A1B6GMH9 A0A1B6MFR0 A0A0A9WPS7
A0A1S3DMK8 A0A023F096 A0A0A9W266 A0A0A9VTB0 A0A1S3DIZ2 A0A0A9VUS3 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A1B6M7K2 A0A0A9X9A3 J9JUM9 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6LLT3 A0A1B6MT91 V5GC07 A0A1S3DMQ9 A0A1S3DM22 A0A3Q0JFQ3 A0A3Q0J0P0 V5GHM7 A0A182I0G9 A0A1S3DMT1 A0A1B6KIX6 A0A3Q0JHG9 A0A3Q0JJJ7 A0A146M0N4 A0A3Q0JBZ9 A0A3Q0J7K9 A0A3Q0J7N1 A0A1Y1LM33 A0A0A9XZV3 A0A1Y1LIG5 A0A0J7K8B6 D6WY83 W8BU98 A0A0J7K7U0 A0A2S2NBR8 A0A1B6GGX5 J9JNP4 A0A2S2QTX5 A0A182WES7 A0A146KZL4 X1X9U0 A0A3Q0IZW4 A0A0J7N391 A0A1S3DBK1 A0A0J7KGQ6 A0A1S3DQE7 W8AH72 A0A0J7N465 A0A0A9Z0W7 X1WPD9 A0A0A9WWW3 A0A2S2NNC8 A0A1B6MIG8 X1XET3 A0A0J7N5Z0 A0A0J7KH73 A0A224XGH3 A0A0A9WHD6 A0A1B6MUZ9 J9JUK4 A0A146KRI6 A0A0J7K952 A0A0J7K9I5 D7EKN2 A0A0J7KNK6 A0A1Y1LBZ3 X1WVF9 J9K0A1 A0A1B6KZV2 A0A2S2NDE3 A0A2S2QWC4 A0A2S2NHU0 A0A0N0PAG9 X1XU26 A0A0V0Z0L1 A0A0A9W8L8 X1X8C2 A0A2S2PT56 X1XTW5 A0A0A9VYL4 A0A0A9YD88 A0A0J7MYK0 A0A146LBU6 A0A0A9VSF7 A0A0J7KC70 X1XB32 A0A1B6GMH9 A0A1B6MFR0 A0A0A9WPS7
EMBL
GAIX01013989
JAA78571.1
GEZM01054789
JAV73399.1
GBBI01004276
JAC14436.1
+ More
GBHO01044644 JAF98959.1 GBHO01044645 JAF98958.1 GBHO01044643 JAF98960.1 GEBQ01008079 JAT31898.1 GBHO01027393 JAG16211.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 GEBQ01015458 JAT24519.1 GEBQ01000825 JAT39152.1 GALX01000776 JAB67690.1 GALX01004947 JAB63519.1 APCN01002077 GEBQ01028569 JAT11408.1 GDHC01006469 JAQ12160.1 GEZM01054766 GEZM01054765 JAV73410.1 GBHO01020799 JAG22805.1 JAV73413.1 LBMM01011720 KMQ86668.1 KQ971357 EFA07702.1 GAMC01009674 JAB96881.1 LBMM01012238 KMQ86334.1 GGMR01002024 MBY14643.1 GECZ01008071 JAS61698.1 ABLF02005684 GGMS01011369 MBY80572.1 GDHC01018192 JAQ00437.1 ABLF02025690 LBMM01010886 KMQ87175.1 LBMM01007770 KMQ89434.1 GAMC01021577 GAMC01021573 JAB84982.1 LBMM01010417 KMQ87475.1 GBHO01005525 JAG38079.1 ABLF02006985 GBHO01034255 GBHO01034254 JAG09349.1 JAG09350.1 GGMR01005823 MBY18442.1 GEBQ01004229 JAT35748.1 ABLF02041762 LBMM01009496 KMQ88090.1 LBMM01007366 KMQ89798.1 GFTR01008866 JAW07560.1 GBHO01037676 JAG05928.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 ABLF02039606 GDHC01019528 JAP99100.1 LBMM01011245 KMQ86943.1 LBMM01011244 KMQ86944.1 DS497760 EFA13225.1 LBMM01005079 KMQ91831.1 GEZM01060254 JAV71154.1 ABLF02026240 ABLF02055027 ABLF02067195 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 GGMR01002187 MBY14806.1 GGMS01012864 MBY82067.1 GGMR01003717 MBY16336.1 KQ458473 KPJ05885.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 JYDQ01000959 KRY06068.1 GBHO01039838 GBHO01006034 JAG03766.1 JAG37570.1 GGMR01019938 MBY32557.1 ABLF02018317 GBHO01044316 JAF99287.1 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 LBMM01013710 KMQ85515.1 GDHC01014057 JAQ04572.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 LBMM01009863 KMQ87831.1 ABLF02056547 GECZ01006169 JAS63600.1 GEBQ01005223 JAT34754.1 GBHO01033147 JAG10457.1
GBHO01044644 JAF98959.1 GBHO01044645 JAF98958.1 GBHO01044643 JAF98960.1 GEBQ01008079 JAT31898.1 GBHO01027393 JAG16211.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 GEBQ01015458 JAT24519.1 GEBQ01000825 JAT39152.1 GALX01000776 JAB67690.1 GALX01004947 JAB63519.1 APCN01002077 GEBQ01028569 JAT11408.1 GDHC01006469 JAQ12160.1 GEZM01054766 GEZM01054765 JAV73410.1 GBHO01020799 JAG22805.1 JAV73413.1 LBMM01011720 KMQ86668.1 KQ971357 EFA07702.1 GAMC01009674 JAB96881.1 LBMM01012238 KMQ86334.1 GGMR01002024 MBY14643.1 GECZ01008071 JAS61698.1 ABLF02005684 GGMS01011369 MBY80572.1 GDHC01018192 JAQ00437.1 ABLF02025690 LBMM01010886 KMQ87175.1 LBMM01007770 KMQ89434.1 GAMC01021577 GAMC01021573 JAB84982.1 LBMM01010417 KMQ87475.1 GBHO01005525 JAG38079.1 ABLF02006985 GBHO01034255 GBHO01034254 JAG09349.1 JAG09350.1 GGMR01005823 MBY18442.1 GEBQ01004229 JAT35748.1 ABLF02041762 LBMM01009496 KMQ88090.1 LBMM01007366 KMQ89798.1 GFTR01008866 JAW07560.1 GBHO01037676 JAG05928.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 ABLF02039606 GDHC01019528 JAP99100.1 LBMM01011245 KMQ86943.1 LBMM01011244 KMQ86944.1 DS497760 EFA13225.1 LBMM01005079 KMQ91831.1 GEZM01060254 JAV71154.1 ABLF02026240 ABLF02055027 ABLF02067195 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 GGMR01002187 MBY14806.1 GGMS01012864 MBY82067.1 GGMR01003717 MBY16336.1 KQ458473 KPJ05885.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 JYDQ01000959 KRY06068.1 GBHO01039838 GBHO01006034 JAG03766.1 JAG37570.1 GGMR01019938 MBY32557.1 ABLF02018317 GBHO01044316 JAF99287.1 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 LBMM01013710 KMQ85515.1 GDHC01014057 JAQ04572.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 LBMM01009863 KMQ87831.1 ABLF02056547 GECZ01006169 JAS63600.1 GEBQ01005223 JAT34754.1 GBHO01033147 JAG10457.1
Proteomes
Pfam
Interpro
IPR008042
Retrotrans_Pao
+ More
IPR041588 Integrase_H2C2
IPR012337 RNaseH-like_sf
IPR040676 DUF5641
IPR005312 DUF1759
IPR036397 RNaseH_sf
IPR008737 Peptidase_asp_put
IPR001584 Integrase_cat-core
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR017996 Royal_jelly/protein_yellow
IPR011042 6-blade_b-propeller_TolB-like
IPR029846 Yellow
IPR001995 Peptidase_A2_cat
IPR038563 Endonuclease_7_sf
IPR004211 Endonuclease_7
IPR001969 Aspartic_peptidase_AS
IPR000727 T_SNARE_dom
IPR015416 Znf_H2C2_histone_UAS-bd
IPR041588 Integrase_H2C2
IPR012337 RNaseH-like_sf
IPR040676 DUF5641
IPR005312 DUF1759
IPR036397 RNaseH_sf
IPR008737 Peptidase_asp_put
IPR001584 Integrase_cat-core
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR017996 Royal_jelly/protein_yellow
IPR011042 6-blade_b-propeller_TolB-like
IPR029846 Yellow
IPR001995 Peptidase_A2_cat
IPR038563 Endonuclease_7_sf
IPR004211 Endonuclease_7
IPR001969 Aspartic_peptidase_AS
IPR000727 T_SNARE_dom
IPR015416 Znf_H2C2_histone_UAS-bd
Gene 3D
ProteinModelPortal
A0A1S3DJC4
S4NIN9
A0A1S3DBQ3
A0A1S3DTS8
A0A1Y1LIC8
A0A1S4EIX5
+ More
A0A1S3DMK8 A0A023F096 A0A0A9W266 A0A0A9VTB0 A0A1S3DIZ2 A0A0A9VUS3 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A1B6M7K2 A0A0A9X9A3 J9JUM9 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6LLT3 A0A1B6MT91 V5GC07 A0A1S3DMQ9 A0A1S3DM22 A0A3Q0JFQ3 A0A3Q0J0P0 V5GHM7 A0A182I0G9 A0A1S3DMT1 A0A1B6KIX6 A0A3Q0JHG9 A0A3Q0JJJ7 A0A146M0N4 A0A3Q0JBZ9 A0A3Q0J7K9 A0A3Q0J7N1 A0A1Y1LM33 A0A0A9XZV3 A0A1Y1LIG5 A0A0J7K8B6 D6WY83 W8BU98 A0A0J7K7U0 A0A2S2NBR8 A0A1B6GGX5 J9JNP4 A0A2S2QTX5 A0A182WES7 A0A146KZL4 X1X9U0 A0A3Q0IZW4 A0A0J7N391 A0A1S3DBK1 A0A0J7KGQ6 A0A1S3DQE7 W8AH72 A0A0J7N465 A0A0A9Z0W7 X1WPD9 A0A0A9WWW3 A0A2S2NNC8 A0A1B6MIG8 X1XET3 A0A0J7N5Z0 A0A0J7KH73 A0A224XGH3 A0A0A9WHD6 A0A1B6MUZ9 J9JUK4 A0A146KRI6 A0A0J7K952 A0A0J7K9I5 D7EKN2 A0A0J7KNK6 A0A1Y1LBZ3 X1WVF9 J9K0A1 A0A1B6KZV2 A0A2S2NDE3 A0A2S2QWC4 A0A2S2NHU0 A0A0N0PAG9 X1XU26 A0A0V0Z0L1 A0A0A9W8L8 X1X8C2 A0A2S2PT56 X1XTW5 A0A0A9VYL4 A0A0A9YD88 A0A0J7MYK0 A0A146LBU6 A0A0A9VSF7 A0A0J7KC70 X1XB32 A0A1B6GMH9 A0A1B6MFR0 A0A0A9WPS7
A0A1S3DMK8 A0A023F096 A0A0A9W266 A0A0A9VTB0 A0A1S3DIZ2 A0A0A9VUS3 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A1B6M7K2 A0A0A9X9A3 J9JUM9 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6LLT3 A0A1B6MT91 V5GC07 A0A1S3DMQ9 A0A1S3DM22 A0A3Q0JFQ3 A0A3Q0J0P0 V5GHM7 A0A182I0G9 A0A1S3DMT1 A0A1B6KIX6 A0A3Q0JHG9 A0A3Q0JJJ7 A0A146M0N4 A0A3Q0JBZ9 A0A3Q0J7K9 A0A3Q0J7N1 A0A1Y1LM33 A0A0A9XZV3 A0A1Y1LIG5 A0A0J7K8B6 D6WY83 W8BU98 A0A0J7K7U0 A0A2S2NBR8 A0A1B6GGX5 J9JNP4 A0A2S2QTX5 A0A182WES7 A0A146KZL4 X1X9U0 A0A3Q0IZW4 A0A0J7N391 A0A1S3DBK1 A0A0J7KGQ6 A0A1S3DQE7 W8AH72 A0A0J7N465 A0A0A9Z0W7 X1WPD9 A0A0A9WWW3 A0A2S2NNC8 A0A1B6MIG8 X1XET3 A0A0J7N5Z0 A0A0J7KH73 A0A224XGH3 A0A0A9WHD6 A0A1B6MUZ9 J9JUK4 A0A146KRI6 A0A0J7K952 A0A0J7K9I5 D7EKN2 A0A0J7KNK6 A0A1Y1LBZ3 X1WVF9 J9K0A1 A0A1B6KZV2 A0A2S2NDE3 A0A2S2QWC4 A0A2S2NHU0 A0A0N0PAG9 X1XU26 A0A0V0Z0L1 A0A0A9W8L8 X1X8C2 A0A2S2PT56 X1XTW5 A0A0A9VYL4 A0A0A9YD88 A0A0J7MYK0 A0A146LBU6 A0A0A9VSF7 A0A0J7KC70 X1XB32 A0A1B6GMH9 A0A1B6MFR0 A0A0A9WPS7
Ontologies
PANTHER
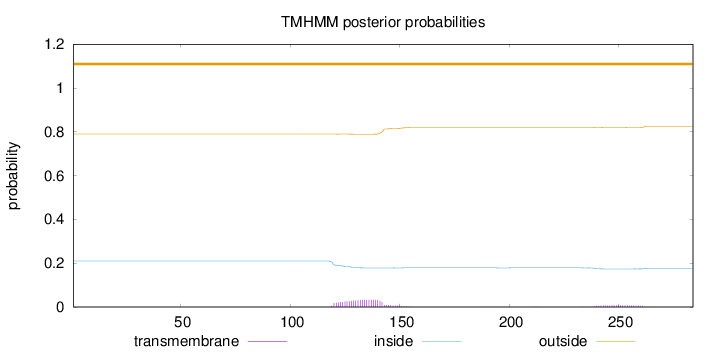
Topology
Length:
284
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.869549999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.21012
outside
1 - 284
Population Genetic Test Statistics
Pi
202.484281
Theta
68.231601
Tajima's D
3.225188
CLR
5.102393
CSRT
0.987050647467627
Interpretation
Uncertain
