Gene
KWMTBOMO15395
Annotation
PREDICTED:_hypoxia-inducible_factor_1-alpha-like_[Bombyx_mori]
Full name
Protein similar
Transcription factor
Location in the cell
Nuclear Reliability : 2.264
Sequence
CDS
ATGTCCGCTTACAAGAACAATGAGAAACGCAAGGAGAAGTCGCGTGTCGCGGCCCGGTGCCGTCGCACCAAAGAGATGCAGATCTTTTCGGAGCTCACGGCGGCCCTGCCTGCAAAAAAGGAGGAAGTTGAGCAACTCGACAAAGCGTCCGTCATGCGTCTGGCTATATCGTACCTTCGCGTACGTGATGTAGTCTCGATGCTTCCAGATGTCAACTCGGATCAATCCAAGATGCAGAATCCCGAAGGCTTCGAAGAGTTAGCTTCAGAGCTGTCATACATGAGGGCTTTGGATGGCTTCGTTCTGGTGCTCTCTCAACAAGGAGACATCGTCTACTGTAGTGACAATATAGCCGAACATCTTGGAGTTTCGCAGACGAAGATGATCTCGCTGTACTAA
Protein
MSAYKNNEKRKEKSRVAARCRRTKEMQIFSELTAALPAKKEEVEQLDKASVMRLAISYLRVRDVVSMLPDVNSDQSKMQNPEGFEELASELSYMRALDGFVLVLSQQGDIVYCSDNIAEHLGVSQTKMISLY
Summary
Description
Functions as a transcriptional regulator of the adaptive response to hypoxia. Binds to core DNA sequence 5'-[AG]CGTG-3' within the hypoxia response element (HRE) of target gene promoters.
Subunit
Efficient DNA binding requires dimerization with another bHLH protein. Interacts with Vhl.
Similarity
Belongs to the tetraspanin (TM4SF) family.
Keywords
Activator
Coiled coil
Complete proteome
Cytoplasm
DNA-binding
Nucleus
Reference proteome
Repeat
Transcription
Transcription regulation
Feature
chain Protein similar
Uniprot
I4DRJ3
A0A2A4JPD3
A0A2W1BY39
A0A2A4JQ85
A0A140CTX5
A0A194Q7C3
+ More
A0A3S2NGW4 A0A194RKJ2 A0A212EZ70 D6WMG7 A0A346QUN2 A0A1Y1LZ17 A0A1Y1LUG8 A0A182JM31 B0W9S7 Q16E64 Q17MC1 A0A1S4EXS9 A0A2P8YCX4 A0A1Q3FJP6 A0A1Q3FJB4 A0A182PR07 A0A182G9V7 A0A1Q3FL92 A0A1B0CAF2 A0A182HTZ9 A0A084W762 A0A1W4VUG7 A0A0K8U5K2 A0A154P5J5 Q8MQV4 B7Z0S3 A0A0L7R7C1 A0A182XHE7 A0A182VP20 A0A182LG42 R9QCX3 Q7PRA0 A0A182JXV3 F4WPX1 B3P7Y5 A0A0J7KS15 Q5U0W0 A0A182QSP3 A0A1L8DGH2 A0A2M4APB6 A0A2M4AP99 A0A0A1XHC8 A0A1L8DGP2 A0A2M4AP63 A0A1W4VGE5 A0A1L8DGH6 A0A1L8DGR9 A0A0K8UI14 Q24167 A0A195AWW6 A0A1W4VTP6 A0A0C9RQD5 B9ER22 B4PLS5 A0A3L8D7M0 B7QI82 A0A026WIE6 W8AKS8 A0A0B4LIX2 D7R4L2 A0A2J7PP09 A0A1B6D4M5 A0A182FL36 A0A310SG20 A0A151WR93 A0A034VAI8 A0A1B6C398 A0A1B6MKJ9 A0A158NHX9 A0A195CKS2 A0A1B6M2U3 A0A131XHJ6 A0A195FEY9 Q5I8Q9 A0A182RND7 A0A2A3E4J2 A0A195EG49 A0A182MJS4 A0A088A014 E0W068 B4R1I3 B4HZL9 V5G121 A0A2R4PB61 A0A3B0JFQ1 W5J2R8 A0A0R3NJU3 A0A182TJF9 B4M6J0 B4JES7 V5HIH3 A0A131Y389 A0A182VYM2
A0A3S2NGW4 A0A194RKJ2 A0A212EZ70 D6WMG7 A0A346QUN2 A0A1Y1LZ17 A0A1Y1LUG8 A0A182JM31 B0W9S7 Q16E64 Q17MC1 A0A1S4EXS9 A0A2P8YCX4 A0A1Q3FJP6 A0A1Q3FJB4 A0A182PR07 A0A182G9V7 A0A1Q3FL92 A0A1B0CAF2 A0A182HTZ9 A0A084W762 A0A1W4VUG7 A0A0K8U5K2 A0A154P5J5 Q8MQV4 B7Z0S3 A0A0L7R7C1 A0A182XHE7 A0A182VP20 A0A182LG42 R9QCX3 Q7PRA0 A0A182JXV3 F4WPX1 B3P7Y5 A0A0J7KS15 Q5U0W0 A0A182QSP3 A0A1L8DGH2 A0A2M4APB6 A0A2M4AP99 A0A0A1XHC8 A0A1L8DGP2 A0A2M4AP63 A0A1W4VGE5 A0A1L8DGH6 A0A1L8DGR9 A0A0K8UI14 Q24167 A0A195AWW6 A0A1W4VTP6 A0A0C9RQD5 B9ER22 B4PLS5 A0A3L8D7M0 B7QI82 A0A026WIE6 W8AKS8 A0A0B4LIX2 D7R4L2 A0A2J7PP09 A0A1B6D4M5 A0A182FL36 A0A310SG20 A0A151WR93 A0A034VAI8 A0A1B6C398 A0A1B6MKJ9 A0A158NHX9 A0A195CKS2 A0A1B6M2U3 A0A131XHJ6 A0A195FEY9 Q5I8Q9 A0A182RND7 A0A2A3E4J2 A0A195EG49 A0A182MJS4 A0A088A014 E0W068 B4R1I3 B4HZL9 V5G121 A0A2R4PB61 A0A3B0JFQ1 W5J2R8 A0A0R3NJU3 A0A182TJF9 B4M6J0 B4JES7 V5HIH3 A0A131Y389 A0A182VYM2
Pubmed
22651552
28756777
26354079
22118469
18362917
19820115
+ More
28004739 17510324 29403074 26483478 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20966253 12364791 21719571 17994087 25830018 8682312 9731218 11006129 12215541 17550304 30249741 24508170 24495485 25348373 21347285 28049606 16311910 20566863 29630918 20920257 23761445 15632085 23185243 25765539
28004739 17510324 29403074 26483478 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20966253 12364791 21719571 17994087 25830018 8682312 9731218 11006129 12215541 17550304 30249741 24508170 24495485 25348373 21347285 28049606 16311910 20566863 29630918 20920257 23761445 15632085 23185243 25765539
EMBL
AK405145
BAM20533.1
NWSH01000838
PCG73925.1
KZ149949
PZC76683.1
+ More
PCG73926.1 KU552106 AMH87782.1 KQ459324 KPJ01442.1 RSAL01000107 RVE47261.1 KQ460045 KPJ18057.1 AGBW02011351 OWR46795.1 KQ971343 EFA04586.2 MG011449 AXS00615.1 GEZM01046369 JAV77285.1 GEZM01046370 JAV77284.1 DS231865 EDS40396.1 CH479045 EAT32521.1 CH477207 EAT47828.1 PYGN01000696 PSN42112.1 GFDL01007301 JAV27744.1 GFDL01007335 JAV27710.1 JXUM01049761 JXUM01049762 KQ561619 KXJ77990.1 GFDL01006740 JAV28305.1 AJWK01003743 AJWK01003744 AJWK01003745 AJWK01003746 ATLV01021144 ATLV01021145 ATLV01021146 ATLV01021147 ATLV01021148 ATLV01021149 ATLV01021150 KE525314 KFB46056.1 GDHF01030498 JAI21816.1 KQ434809 KZC06400.1 AY122269 AAM52781.1 AE014297 ACL83585.1 KQ414643 KOC66656.1 JN228344 AFL70631.1 AAAB01008859 EAA07597.6 GL888255 EGI63800.1 CH954182 EDV53112.1 LBMM01003871 KMQ93034.1 BT016132 AAV37017.1 AXCN02000341 GFDF01008526 JAV05558.1 GGFK01009315 MBW42636.1 GGFK01009266 MBW42587.1 GBXI01005514 GBXI01003935 JAD08778.1 JAD10357.1 GFDF01008527 JAV05557.1 GGFK01009255 MBW42576.1 GFDF01008525 JAV05559.1 GFDF01008524 JAV05560.1 GDHF01026111 JAI26203.1 U43090 KQ976725 KYM76547.1 GBYB01009416 JAG79183.1 BT058097 ACM89953.1 CM000160 EDW99062.1 QOIP01000012 RLU16093.1 ABJB010008711 ABJB010089786 ABJB010570512 ABJB010651084 DS944282 EEC18554.1 KK107185 EZA55822.1 GAMC01021252 JAB85303.1 AHN57598.1 HM032914 ADH01740.1 NEVH01023281 PNF18058.1 GEDC01023846 GEDC01016705 JAS13452.1 JAS20593.1 KQ764717 OAD54410.1 KQ982813 KYQ50321.1 GAKP01019418 JAC39534.1 GEDC01029444 JAS07854.1 GEBQ01003554 JAT36423.1 ADTU01016101 ADTU01016102 KQ977622 KYN01325.1 GEBQ01009708 JAT30269.1 GEFH01002916 JAP65665.1 KQ981636 KYN38787.1 AY845427 AAW31092.1 KZ288379 PBC26608.1 KQ978957 KYN27213.1 AXCM01007259 DS235857 EEB19024.1 CM000364 EDX14971.1 CH480819 EDW53476.1 GALX01004756 JAB63710.1 MG552486 AVX48316.1 OUUW01000005 SPP80955.1 ADMH02002130 ETN58582.1 CM000070 KRS99540.1 KRS99541.1 CH940652 EDW59266.2 CH916369 EDV93208.1 GANP01001023 JAB83445.1 GEFM01002966 JAP72830.1
PCG73926.1 KU552106 AMH87782.1 KQ459324 KPJ01442.1 RSAL01000107 RVE47261.1 KQ460045 KPJ18057.1 AGBW02011351 OWR46795.1 KQ971343 EFA04586.2 MG011449 AXS00615.1 GEZM01046369 JAV77285.1 GEZM01046370 JAV77284.1 DS231865 EDS40396.1 CH479045 EAT32521.1 CH477207 EAT47828.1 PYGN01000696 PSN42112.1 GFDL01007301 JAV27744.1 GFDL01007335 JAV27710.1 JXUM01049761 JXUM01049762 KQ561619 KXJ77990.1 GFDL01006740 JAV28305.1 AJWK01003743 AJWK01003744 AJWK01003745 AJWK01003746 ATLV01021144 ATLV01021145 ATLV01021146 ATLV01021147 ATLV01021148 ATLV01021149 ATLV01021150 KE525314 KFB46056.1 GDHF01030498 JAI21816.1 KQ434809 KZC06400.1 AY122269 AAM52781.1 AE014297 ACL83585.1 KQ414643 KOC66656.1 JN228344 AFL70631.1 AAAB01008859 EAA07597.6 GL888255 EGI63800.1 CH954182 EDV53112.1 LBMM01003871 KMQ93034.1 BT016132 AAV37017.1 AXCN02000341 GFDF01008526 JAV05558.1 GGFK01009315 MBW42636.1 GGFK01009266 MBW42587.1 GBXI01005514 GBXI01003935 JAD08778.1 JAD10357.1 GFDF01008527 JAV05557.1 GGFK01009255 MBW42576.1 GFDF01008525 JAV05559.1 GFDF01008524 JAV05560.1 GDHF01026111 JAI26203.1 U43090 KQ976725 KYM76547.1 GBYB01009416 JAG79183.1 BT058097 ACM89953.1 CM000160 EDW99062.1 QOIP01000012 RLU16093.1 ABJB010008711 ABJB010089786 ABJB010570512 ABJB010651084 DS944282 EEC18554.1 KK107185 EZA55822.1 GAMC01021252 JAB85303.1 AHN57598.1 HM032914 ADH01740.1 NEVH01023281 PNF18058.1 GEDC01023846 GEDC01016705 JAS13452.1 JAS20593.1 KQ764717 OAD54410.1 KQ982813 KYQ50321.1 GAKP01019418 JAC39534.1 GEDC01029444 JAS07854.1 GEBQ01003554 JAT36423.1 ADTU01016101 ADTU01016102 KQ977622 KYN01325.1 GEBQ01009708 JAT30269.1 GEFH01002916 JAP65665.1 KQ981636 KYN38787.1 AY845427 AAW31092.1 KZ288379 PBC26608.1 KQ978957 KYN27213.1 AXCM01007259 DS235857 EEB19024.1 CM000364 EDX14971.1 CH480819 EDW53476.1 GALX01004756 JAB63710.1 MG552486 AVX48316.1 OUUW01000005 SPP80955.1 ADMH02002130 ETN58582.1 CM000070 KRS99540.1 KRS99541.1 CH940652 EDW59266.2 CH916369 EDV93208.1 GANP01001023 JAB83445.1 GEFM01002966 JAP72830.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000053240
UP000007151
UP000007266
+ More
UP000075880 UP000002320 UP000008820 UP000245037 UP000075885 UP000069940 UP000249989 UP000092461 UP000030765 UP000192221 UP000076502 UP000000803 UP000053825 UP000076407 UP000075903 UP000075882 UP000007062 UP000075881 UP000007755 UP000008711 UP000036403 UP000075886 UP000078540 UP000002282 UP000279307 UP000001555 UP000053097 UP000235965 UP000069272 UP000075809 UP000005205 UP000078542 UP000078541 UP000075900 UP000242457 UP000078492 UP000075883 UP000005203 UP000009046 UP000000304 UP000001292 UP000268350 UP000000673 UP000001819 UP000075902 UP000008792 UP000001070 UP000075920
UP000075880 UP000002320 UP000008820 UP000245037 UP000075885 UP000069940 UP000249989 UP000092461 UP000030765 UP000192221 UP000076502 UP000000803 UP000053825 UP000076407 UP000075903 UP000075882 UP000007062 UP000075881 UP000007755 UP000008711 UP000036403 UP000075886 UP000078540 UP000002282 UP000279307 UP000001555 UP000053097 UP000235965 UP000069272 UP000075809 UP000005205 UP000078542 UP000078541 UP000075900 UP000242457 UP000078492 UP000075883 UP000005203 UP000009046 UP000000304 UP000001292 UP000268350 UP000000673 UP000001819 UP000075902 UP000008792 UP000001070 UP000075920
Pfam
Interpro
Gene 3D
ProteinModelPortal
I4DRJ3
A0A2A4JPD3
A0A2W1BY39
A0A2A4JQ85
A0A140CTX5
A0A194Q7C3
+ More
A0A3S2NGW4 A0A194RKJ2 A0A212EZ70 D6WMG7 A0A346QUN2 A0A1Y1LZ17 A0A1Y1LUG8 A0A182JM31 B0W9S7 Q16E64 Q17MC1 A0A1S4EXS9 A0A2P8YCX4 A0A1Q3FJP6 A0A1Q3FJB4 A0A182PR07 A0A182G9V7 A0A1Q3FL92 A0A1B0CAF2 A0A182HTZ9 A0A084W762 A0A1W4VUG7 A0A0K8U5K2 A0A154P5J5 Q8MQV4 B7Z0S3 A0A0L7R7C1 A0A182XHE7 A0A182VP20 A0A182LG42 R9QCX3 Q7PRA0 A0A182JXV3 F4WPX1 B3P7Y5 A0A0J7KS15 Q5U0W0 A0A182QSP3 A0A1L8DGH2 A0A2M4APB6 A0A2M4AP99 A0A0A1XHC8 A0A1L8DGP2 A0A2M4AP63 A0A1W4VGE5 A0A1L8DGH6 A0A1L8DGR9 A0A0K8UI14 Q24167 A0A195AWW6 A0A1W4VTP6 A0A0C9RQD5 B9ER22 B4PLS5 A0A3L8D7M0 B7QI82 A0A026WIE6 W8AKS8 A0A0B4LIX2 D7R4L2 A0A2J7PP09 A0A1B6D4M5 A0A182FL36 A0A310SG20 A0A151WR93 A0A034VAI8 A0A1B6C398 A0A1B6MKJ9 A0A158NHX9 A0A195CKS2 A0A1B6M2U3 A0A131XHJ6 A0A195FEY9 Q5I8Q9 A0A182RND7 A0A2A3E4J2 A0A195EG49 A0A182MJS4 A0A088A014 E0W068 B4R1I3 B4HZL9 V5G121 A0A2R4PB61 A0A3B0JFQ1 W5J2R8 A0A0R3NJU3 A0A182TJF9 B4M6J0 B4JES7 V5HIH3 A0A131Y389 A0A182VYM2
A0A3S2NGW4 A0A194RKJ2 A0A212EZ70 D6WMG7 A0A346QUN2 A0A1Y1LZ17 A0A1Y1LUG8 A0A182JM31 B0W9S7 Q16E64 Q17MC1 A0A1S4EXS9 A0A2P8YCX4 A0A1Q3FJP6 A0A1Q3FJB4 A0A182PR07 A0A182G9V7 A0A1Q3FL92 A0A1B0CAF2 A0A182HTZ9 A0A084W762 A0A1W4VUG7 A0A0K8U5K2 A0A154P5J5 Q8MQV4 B7Z0S3 A0A0L7R7C1 A0A182XHE7 A0A182VP20 A0A182LG42 R9QCX3 Q7PRA0 A0A182JXV3 F4WPX1 B3P7Y5 A0A0J7KS15 Q5U0W0 A0A182QSP3 A0A1L8DGH2 A0A2M4APB6 A0A2M4AP99 A0A0A1XHC8 A0A1L8DGP2 A0A2M4AP63 A0A1W4VGE5 A0A1L8DGH6 A0A1L8DGR9 A0A0K8UI14 Q24167 A0A195AWW6 A0A1W4VTP6 A0A0C9RQD5 B9ER22 B4PLS5 A0A3L8D7M0 B7QI82 A0A026WIE6 W8AKS8 A0A0B4LIX2 D7R4L2 A0A2J7PP09 A0A1B6D4M5 A0A182FL36 A0A310SG20 A0A151WR93 A0A034VAI8 A0A1B6C398 A0A1B6MKJ9 A0A158NHX9 A0A195CKS2 A0A1B6M2U3 A0A131XHJ6 A0A195FEY9 Q5I8Q9 A0A182RND7 A0A2A3E4J2 A0A195EG49 A0A182MJS4 A0A088A014 E0W068 B4R1I3 B4HZL9 V5G121 A0A2R4PB61 A0A3B0JFQ1 W5J2R8 A0A0R3NJU3 A0A182TJF9 B4M6J0 B4JES7 V5HIH3 A0A131Y389 A0A182VYM2
PDB
4ZPR
E-value=1.07529e-22,
Score=257
Ontologies
KEGG
PATHWAY
GO
GO:0046983
GO:0005634
GO:0005737
GO:0003700
GO:0005667
GO:0003677
GO:0006355
GO:0000981
GO:0006357
GO:0045944
GO:0060438
GO:0071456
GO:0010508
GO:0030308
GO:0008286
GO:0048477
GO:0090575
GO:0005829
GO:0032869
GO:0030334
GO:0045088
GO:0001228
GO:0003989
GO:0016021
GO:0043565
GO:0008134
GO:0019904
GO:0003697
GO:0008094
GO:0005524
GO:0006281
GO:0006259
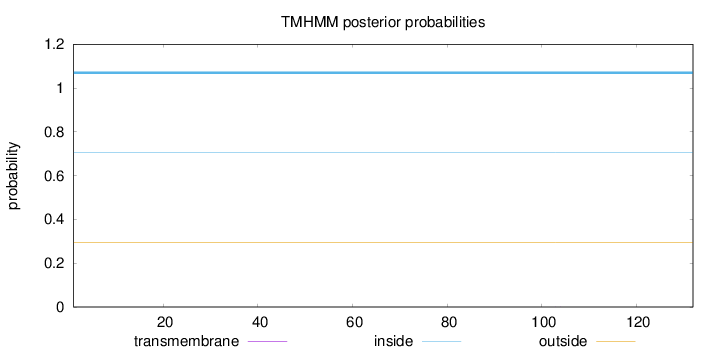
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
Length:
132
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00263
Exp number, first 60 AAs:
0.00012
Total prob of N-in:
0.70563
inside
1 - 132
Population Genetic Test Statistics
Pi
149.980829
Theta
157.892059
Tajima's D
-0.508915
CLR
11.802575
CSRT
0.23938803059847
Interpretation
Uncertain
