Gene
KWMTBOMO15389
Pre Gene Modal
BGIBMGA005102
Annotation
PREDICTED:_chromosome-associated_kinesin_KIF4_[Amyelois_transitella]
Full name
Kinesin-like protein
+ More
Chromosome-associated kinesin KIF4
Chromosome-associated kinesin KIF4
Alternative Name
Chromokinesin
Chromosome-associated kinesin KLP1
Chromosome-associated kinesin KLP1
Location in the cell
Nuclear Reliability : 2.206
Sequence
CDS
ATGGTTGAAGATAAAACAATAGACACTGTTCAGGTAGCGCTGCGCATCAGACCTTTGATGCAACAGGAGACCGAAAGAGGCTGCGATGAGTGCATTGACGTGATACCTGGGGAGCAACAAGTGCAAATAAAAGACTTAGCTTTTACATACAATTACGTATTCGCGCAGCATATCACTCAAAGAGAATTTTATGACACTGCTGTGAAAGGACTAATAGGAAAGTTATTTCAAGGGTACAATGTAACAATACTGGCATATGGCCAAACAGGGTCAGGCAAAACTTATACCATGGGTACTAATTACTCTGGGTCAGATGGAGACTTCACTAAACTTGGAGTAATCCCGCAAGCAGTGGCGGACATCTTTGACTTTATTGAAACTCATGAAGACAAGTTCATATTCAAAGTGAATGTCTCTTTCATGGAGCTATACCAGGAGCAGTGCTACGACCTGTTGTCAGGGAAGGAGCGTGGACACAGTATTATCGAGATCAGAGAGGATATCAATAAAGGTGTAGTTTTGCCAGGTATAACCGAATTACCTGTGGCATCCACAATGGAGACAATGCTAGTATTAGAACGAGGGTCTGCTGGTCGTGTAACAGGATCGACAGCAATGAACCAAGCATCCAGTCGAAGTCATGCCGTTTTTACAATTATAATTTGTAAAGAGAGTCGAAGTGATAAAAATATAGCAACAACATCTAAGTTTCATCTCGTCGATTTGGCTGGTTCAGAACGCATAAAAAAGACGAAAGCTAGTGGAGAGCGATTGAAAGAAGGTGTGAAGATCAATCAAGGTCTACTTGCACTTGGCAACGTAATTTCCGCTCTCGGAGATGGAACCAATAGAAGTTATATCAGTTACAGAGATAGCAAGCTCACTAGGCTTCTTCAGGACAGTCTGGGAGGCAACTCGCTGACGCTGATGGTGGCGTGTGTGAGTCCGGCCGACTACAACCTAGACGAGACCGTGTCCACGCTGCGCTACGCCGACCGCGCTCGGCGGATCCGGAACAAACCGGTCATCAACCAGGACGCTAAGGCCGCCGAGATCGTAAGGTTAAACAATTTAGTCAACGAACTCCGACTGCAGCTTTTAGGCAAATTGCCAACTGTTAATGAACAAAATAATGAAAAACTGCAAGAGGAGTTAGAAAGGGAACGATCCAGATATGCAGAGCTGTTGAAGAAACACAAACAAGTCACTGAACATCTTGGCAACATGTTAATTGAGAATACAAATTTATGCGAAAAAGCACTTTTAGCTGAAGCAGCCAAGGACAAAATAGAACGCAAACTGAACGAGCTCACGGAACAGTGCAATCAGACCATTGAAAATCTTAACATGACTGATAAATCTGATGATGAGCACCAGAAGACATCAGTAGTCGATTATTTGAAAGAAATTAAGACTAGGCTTGAAGACTTGCAGTCTGTCAACTTGAAGAACAATGAGGAATTAATTGATCATGAAATCAAGCTATCGTTTGTCAAAGATGACAATGATCATGAGAAAGTTGATGAAGATGTCGTATTAAATGAGGATCAGGCAGTTTTGGAAGAAGAGAAAAGGGCAATGGGACAGGTGGCACTAAATCAAGAATTACAAGAATTAAACCGTGCCATGGCCATTAAGGCGTCAGTAGTACAAGCAATATTAGCCAACAATAAAGAAATGCTGGACTCCCATCATAATTTACGTGAAAACGAGGAAAAGATCTCGCACCTAGAGAAAGAACGAGATGAGTTACTTCAGCAATTGAAACAGACTAAGAGCAAAGATCCATCGCACGAAGAGAGACGCACGAAAGTATCGACCCTCGAATCTGAGATATCTGAACTGAAGAAGAGATGCCAGCAGCAAGCAAACATCATCAAGATGAAAGAGAAAAACGAAGCTAAGATAGCATCACTCAATGCTGAACTGCAAGCTATGAAAGTTACTAAGGTTAAAATAATAAAACAAATGCGCGAAGAAAGCGAGAAGTTCCGTAAGTGGAAGGCGGACAACGAGCGCGCCCTGCTGCGGCTCAGGAACGAGGATCGGAAGCGCGCCACCGCCATGGCCAAGATGGAGACTCTGCACGCTAAACAACAGAACGTACTGAAACGGAAGATGGAAGAGGCTGTCGCAGTTAACAGAAGACTTAAGGAAGCATTGGATCGTCAGAAGACGACCGCAATGAAGCGCAGCGCTAAAGGCAACGTGAAGGCGGGCGCCGTGCAGCAGTACATCGAACAAGAACTCGAAGTTCATCTCAGTATTGTCGAAGCCGAGAAGTCCTTGGAAGAACTCATGGAGTACAGGGCTTGGATCACGGAACAAATCGAGAGTCTTCGCAACAGTACAGACTCGGAGGTCAACAAAAAGAAAATTGCTGAACTTGAGGATGACCTCGCTCTGCGTAAGGCTCAGATATCAGATTTGCAGCAGAAGATATTGACAGCAGATCAAGAAAATAAATCACGCACGCAATGGGACAACATACAGTCCATGCTCGAAGCGAAGGTCGCTCTCAAATGTCTTTTCGAGTTGCTGGTCGACGCCAAGAGAGAGCTGCAGAACCAGAGCGAGAAGGGCTACCAGGCACGATACGAGGAGGTCAAGGAGGCCCACGACCGGCTGGCCGTCGAGTTCGACAACAGCAAGGCGGAATTCGAAAGGCAGCTGATGAATGTCAAGAAACAGAGCGAACAAAAGCTGACGGCGCTACTGGCGCTGCAGCGCGGCGTGGTGCGCGGCGACAAGGGCGAGGCCTGCAAGCATCTGCACAACGTCATACAGGCGCAGCAGGACAAGCTCGAGCAAGTTGAAGATGAAAACAAAAAACTGATGGAAGAATTAGAAGAGCTACGTGCGGCCGGCAAACGCGCCAAGAAACGGTCGTCGAAGAAGGAGTCCTCGGGCGAGGTCAAGAAACAGGAGTACACCGAGCTCACTGATGAGGACGACGACGAGTTTGAGGACAGAGACAAGGACCCAGACTGGAGGGCCACGCCTCTGTTCAAACGGATACAGATCAAAAAAAAAATTTAA
Protein
MVEDKTIDTVQVALRIRPLMQQETERGCDECIDVIPGEQQVQIKDLAFTYNYVFAQHITQREFYDTAVKGLIGKLFQGYNVTILAYGQTGSGKTYTMGTNYSGSDGDFTKLGVIPQAVADIFDFIETHEDKFIFKVNVSFMELYQEQCYDLLSGKERGHSIIEIREDINKGVVLPGITELPVASTMETMLVLERGSAGRVTGSTAMNQASSRSHAVFTIIICKESRSDKNIATTSKFHLVDLAGSERIKKTKASGERLKEGVKINQGLLALGNVISALGDGTNRSYISYRDSKLTRLLQDSLGGNSLTLMVACVSPADYNLDETVSTLRYADRARRIRNKPVINQDAKAAEIVRLNNLVNELRLQLLGKLPTVNEQNNEKLQEELERERSRYAELLKKHKQVTEHLGNMLIENTNLCEKALLAEAAKDKIERKLNELTEQCNQTIENLNMTDKSDDEHQKTSVVDYLKEIKTRLEDLQSVNLKNNEELIDHEIKLSFVKDDNDHEKVDEDVVLNEDQAVLEEEKRAMGQVALNQELQELNRAMAIKASVVQAILANNKEMLDSHHNLRENEEKISHLEKERDELLQQLKQTKSKDPSHEERRTKVSTLESEISELKKRCQQQANIIKMKEKNEAKIASLNAELQAMKVTKVKIIKQMREESEKFRKWKADNERALLRLRNEDRKRATAMAKMETLHAKQQNVLKRKMEEAVAVNRRLKEALDRQKTTAMKRSAKGNVKAGAVQQYIEQELEVHLSIVEAEKSLEELMEYRAWITEQIESLRNSTDSEVNKKKIAELEDDLALRKAQISDLQQKILTADQENKSRTQWDNIQSMLEAKVALKCLFELLVDAKRELQNQSEKGYQARYEEVKEAHDRLAVEFDNSKAEFERQLMNVKKQSEQKLTALLALQRGVVRGDKGEACKHLHNVIQAQQDKLEQVEDENKKLMEELEELRAAGKRAKKRSSKKESSGEVKKQEYTELTDEDDDEFEDRDKDPDWRATPLFKRIQIKKKI
Summary
Description
Required for mitotic chromosomal positioning and bipolar spindle stabilization.
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. Chromokinesin subfamily.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. Chromokinesin subfamily.
Keywords
ATP-binding
Chromosome
Coiled coil
Cytoplasm
Cytoskeleton
DNA-binding
Microtubule
Motor protein
Nucleotide-binding
Nucleus
Feature
chain Chromosome-associated kinesin KIF4
Uniprot
H9J6G0
A0A2A4JUJ3
A0A2W1BV73
A0A2H1WQ29
A0A194RKI6
A0A194Q7D3
+ More
A0A2A4JVP0 A0A3S2LYT7 A0A194Q908 A0A212EJ78 A0A0N1INR7 A0A195FK11 A0A158P2Q9 A0A151I0F6 F4W5R8 A0A195EZL8 F4WXB4 A0A151X2K6 A0A2A3E9X3 A0A195DFE9 A0A088A2C5 A0A0N0BGY7 A0A195C3L1 E2AZU7 A0A1Y1MH00 A0A154P8G1 A0A026VV16 A0A3L8D4X3 D6WTV8 E9JC95 A0A1W4XDT4 A0A1W4XPH6 A0A1W4XE54 E0VBK4 E2BRZ7 A0A1B6E537 A0A2P8Y0R4 K7IQ94 A0A232EX98 T1PMI9 A0A1I8MRE6 A0A1I8MRG6 A0A0L7QXV6 A0A1I8MRE0 A0A0C9RNS9 A0A0J7MYN5 A0A1B6JEE1 A0A1B6JQV7 N6TLR8 U4UF76 A0A1A9YN98 A0A1Q3EWD5 A0A1B0C5I3 A0A1A9ZHG6 A0A2G8KPF9 A0A0K8S753 A0A0A9YB23 A0A023F1R6 A0A146LR33 A0A1B0FEL6 J9K176 A0A1S3KAT7 A0A2H8TQC5 A0A1S3IN85 A0A1A9VPB0 A0A023EZD1 A0A2S2NYZ2 H0YWL1 A0A3B3S2J6 Q91784 Q6IRM2 A0A3B3S4F8 H9G9C3 A0A2S2Q0P6 A0A2P4TAY7 A0A226N556 A0A0L7R797 F6ZGY3 F6TNW7 A0A093QU19 A0A2I0LS47 G1MS31 A0A3L8SIC2 A0A1S3A805 A0A091L5T7 V9K8S5 A0A0A0A7J7 E9PSJ3 M3W9X8 A0A384DUB0 A0A3Q7VUB6 A0A1U7RSD0 A0A091V288 W5Q1N4 A0A1S3GE59 D2H8A0 A0A1S3GE90
A0A2A4JVP0 A0A3S2LYT7 A0A194Q908 A0A212EJ78 A0A0N1INR7 A0A195FK11 A0A158P2Q9 A0A151I0F6 F4W5R8 A0A195EZL8 F4WXB4 A0A151X2K6 A0A2A3E9X3 A0A195DFE9 A0A088A2C5 A0A0N0BGY7 A0A195C3L1 E2AZU7 A0A1Y1MH00 A0A154P8G1 A0A026VV16 A0A3L8D4X3 D6WTV8 E9JC95 A0A1W4XDT4 A0A1W4XPH6 A0A1W4XE54 E0VBK4 E2BRZ7 A0A1B6E537 A0A2P8Y0R4 K7IQ94 A0A232EX98 T1PMI9 A0A1I8MRE6 A0A1I8MRG6 A0A0L7QXV6 A0A1I8MRE0 A0A0C9RNS9 A0A0J7MYN5 A0A1B6JEE1 A0A1B6JQV7 N6TLR8 U4UF76 A0A1A9YN98 A0A1Q3EWD5 A0A1B0C5I3 A0A1A9ZHG6 A0A2G8KPF9 A0A0K8S753 A0A0A9YB23 A0A023F1R6 A0A146LR33 A0A1B0FEL6 J9K176 A0A1S3KAT7 A0A2H8TQC5 A0A1S3IN85 A0A1A9VPB0 A0A023EZD1 A0A2S2NYZ2 H0YWL1 A0A3B3S2J6 Q91784 Q6IRM2 A0A3B3S4F8 H9G9C3 A0A2S2Q0P6 A0A2P4TAY7 A0A226N556 A0A0L7R797 F6ZGY3 F6TNW7 A0A093QU19 A0A2I0LS47 G1MS31 A0A3L8SIC2 A0A1S3A805 A0A091L5T7 V9K8S5 A0A0A0A7J7 E9PSJ3 M3W9X8 A0A384DUB0 A0A3Q7VUB6 A0A1U7RSD0 A0A091V288 W5Q1N4 A0A1S3GE59 D2H8A0 A0A1S3GE90
Pubmed
19121390
28756777
26354079
22118469
21347285
21719571
+ More
20798317 28004739 24508170 30249741 18362917 19820115 21282665 20566863 29403074 20075255 28648823 25315136 23537049 29023486 25401762 26823975 25474469 20360741 29240929 7720067 8482413 27762356 21881562 19892987 17495919 23371554 20838655 30282656 24402279 15057822 22673903 17975172 20809919 20010809
20798317 28004739 24508170 30249741 18362917 19820115 21282665 20566863 29403074 20075255 28648823 25315136 23537049 29023486 25401762 26823975 25474469 20360741 29240929 7720067 8482413 27762356 21881562 19892987 17495919 23371554 20838655 30282656 24402279 15057822 22673903 17975172 20809919 20010809
EMBL
BABH01019665
NWSH01000553
PCG75695.1
KZ149949
PZC76690.1
ODYU01010176
+ More
SOQ55117.1 KQ460045 KPJ18052.1 KQ459324 KPJ01452.1 PCG75694.1 RSAL01000107 RVE47268.1 KPJ01450.1 AGBW02014519 OWR41535.1 KQ461045 KPJ09949.1 KQ981522 KYN40691.1 ADTU01007389 ADTU01007390 KQ976609 KYM79308.1 GL887695 EGI70394.1 KQ981905 KYN33324.1 GL888423 EGI61163.1 KQ982576 KYQ54677.1 KZ288311 PBC28525.1 KQ980903 KYN11586.1 KQ435762 KOX75484.1 KQ978344 KYM94776.1 GL444277 EFN61076.1 GEZM01036563 JAV82587.1 KQ434844 KZC08147.1 KK107796 EZA47603.1 QOIP01000013 RLU15547.1 KQ971352 EFA07572.2 GL771776 EFZ09561.1 DS235032 EEB10760.1 GL450107 EFN81513.1 GEDC01004257 JAS33041.1 PYGN01001074 PSN37848.1 NNAY01001780 OXU22938.1 KA649996 AFP64625.1 KQ414700 KOC63386.1 GBYB01010089 GBYB01010090 JAG79856.1 JAG79857.1 LBMM01013627 KMQ85555.1 GECU01010228 JAS97478.1 GECU01006082 JAT01625.1 APGK01019272 APGK01019273 KB740098 ENN81394.1 KB632331 ERL92629.1 GFDL01015430 JAV19615.1 JXJN01026022 MRZV01000445 PIK49840.1 GBRD01017277 JAG48550.1 GBHO01013327 GBRD01017276 GDHC01010443 JAG30277.1 JAG48551.1 JAQ08186.1 GBBI01003766 JAC14946.1 GDHC01009482 JAQ09147.1 CCAG010008349 ABLF02012299 GFXV01004027 MBW15832.1 GBBI01004115 JAC14597.1 GGMR01009830 MBY22449.1 ABQF01018162 X82012 BC070854 CM004481 AAH70854.1 OCT65724.1 AAWZ02027079 GGMS01002066 MBY71269.1 PPHD01003389 POI33532.1 MCFN01000203 OXB62671.1 KQ414642 KOC66714.1 KL424336 KFW89870.1 AKCR02000116 PKK20257.1 QUSF01000018 RLW02571.1 KL297335 KFP50618.1 JW861575 AFO94092.1 KL871140 KGL90584.1 AABR07073536 AC141377 AANG04000501 KL410570 KFQ97119.1 AMGL01115865 AMGL01115866 AMGL01115867 AMGL01115868 AMGL01115869 AMGL01115870 AMGL01115871 AMGL01115872 GL192572 EFB28133.1
SOQ55117.1 KQ460045 KPJ18052.1 KQ459324 KPJ01452.1 PCG75694.1 RSAL01000107 RVE47268.1 KPJ01450.1 AGBW02014519 OWR41535.1 KQ461045 KPJ09949.1 KQ981522 KYN40691.1 ADTU01007389 ADTU01007390 KQ976609 KYM79308.1 GL887695 EGI70394.1 KQ981905 KYN33324.1 GL888423 EGI61163.1 KQ982576 KYQ54677.1 KZ288311 PBC28525.1 KQ980903 KYN11586.1 KQ435762 KOX75484.1 KQ978344 KYM94776.1 GL444277 EFN61076.1 GEZM01036563 JAV82587.1 KQ434844 KZC08147.1 KK107796 EZA47603.1 QOIP01000013 RLU15547.1 KQ971352 EFA07572.2 GL771776 EFZ09561.1 DS235032 EEB10760.1 GL450107 EFN81513.1 GEDC01004257 JAS33041.1 PYGN01001074 PSN37848.1 NNAY01001780 OXU22938.1 KA649996 AFP64625.1 KQ414700 KOC63386.1 GBYB01010089 GBYB01010090 JAG79856.1 JAG79857.1 LBMM01013627 KMQ85555.1 GECU01010228 JAS97478.1 GECU01006082 JAT01625.1 APGK01019272 APGK01019273 KB740098 ENN81394.1 KB632331 ERL92629.1 GFDL01015430 JAV19615.1 JXJN01026022 MRZV01000445 PIK49840.1 GBRD01017277 JAG48550.1 GBHO01013327 GBRD01017276 GDHC01010443 JAG30277.1 JAG48551.1 JAQ08186.1 GBBI01003766 JAC14946.1 GDHC01009482 JAQ09147.1 CCAG010008349 ABLF02012299 GFXV01004027 MBW15832.1 GBBI01004115 JAC14597.1 GGMR01009830 MBY22449.1 ABQF01018162 X82012 BC070854 CM004481 AAH70854.1 OCT65724.1 AAWZ02027079 GGMS01002066 MBY71269.1 PPHD01003389 POI33532.1 MCFN01000203 OXB62671.1 KQ414642 KOC66714.1 KL424336 KFW89870.1 AKCR02000116 PKK20257.1 QUSF01000018 RLW02571.1 KL297335 KFP50618.1 JW861575 AFO94092.1 KL871140 KGL90584.1 AABR07073536 AC141377 AANG04000501 KL410570 KFQ97119.1 AMGL01115865 AMGL01115866 AMGL01115867 AMGL01115868 AMGL01115869 AMGL01115870 AMGL01115871 AMGL01115872 GL192572 EFB28133.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000078541 UP000005205 UP000078540 UP000007755 UP000075809 UP000242457 UP000078492 UP000005203 UP000053105 UP000078542 UP000000311 UP000076502 UP000053097 UP000279307 UP000007266 UP000192223 UP000009046 UP000008237 UP000245037 UP000002358 UP000215335 UP000095301 UP000053825 UP000036403 UP000019118 UP000030742 UP000092443 UP000092460 UP000092445 UP000230750 UP000092444 UP000007819 UP000085678 UP000078200 UP000007754 UP000261540 UP000186698 UP000001646 UP000198323 UP000002281 UP000002280 UP000053872 UP000001645 UP000276834 UP000079721 UP000053858 UP000002494 UP000011712 UP000261680 UP000286642 UP000189705 UP000053283 UP000002356 UP000081671
UP000078541 UP000005205 UP000078540 UP000007755 UP000075809 UP000242457 UP000078492 UP000005203 UP000053105 UP000078542 UP000000311 UP000076502 UP000053097 UP000279307 UP000007266 UP000192223 UP000009046 UP000008237 UP000245037 UP000002358 UP000215335 UP000095301 UP000053825 UP000036403 UP000019118 UP000030742 UP000092443 UP000092460 UP000092445 UP000230750 UP000092444 UP000007819 UP000085678 UP000078200 UP000007754 UP000261540 UP000186698 UP000001646 UP000198323 UP000002281 UP000002280 UP000053872 UP000001645 UP000276834 UP000079721 UP000053858 UP000002494 UP000011712 UP000261680 UP000286642 UP000189705 UP000053283 UP000002356 UP000081671
Pfam
Interpro
IPR027417
P-loop_NTPase
+ More
IPR019821 Kinesin_motor_CS
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR033467 Tesmin/TSO1-like_CXC
IPR005172 CRC
IPR005744 Hy-lIII
IPR004254 AdipoR/HlyIII-related
IPR004240 EMP70
IPR012943 Cnn_1N
IPR013087 Znf_C2H2_type
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
IPR019821 Kinesin_motor_CS
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR033467 Tesmin/TSO1-like_CXC
IPR005172 CRC
IPR005744 Hy-lIII
IPR004254 AdipoR/HlyIII-related
IPR004240 EMP70
IPR012943 Cnn_1N
IPR013087 Znf_C2H2_type
IPR012934 Znf_AD
IPR036236 Znf_C2H2_sf
Gene 3D
ProteinModelPortal
H9J6G0
A0A2A4JUJ3
A0A2W1BV73
A0A2H1WQ29
A0A194RKI6
A0A194Q7D3
+ More
A0A2A4JVP0 A0A3S2LYT7 A0A194Q908 A0A212EJ78 A0A0N1INR7 A0A195FK11 A0A158P2Q9 A0A151I0F6 F4W5R8 A0A195EZL8 F4WXB4 A0A151X2K6 A0A2A3E9X3 A0A195DFE9 A0A088A2C5 A0A0N0BGY7 A0A195C3L1 E2AZU7 A0A1Y1MH00 A0A154P8G1 A0A026VV16 A0A3L8D4X3 D6WTV8 E9JC95 A0A1W4XDT4 A0A1W4XPH6 A0A1W4XE54 E0VBK4 E2BRZ7 A0A1B6E537 A0A2P8Y0R4 K7IQ94 A0A232EX98 T1PMI9 A0A1I8MRE6 A0A1I8MRG6 A0A0L7QXV6 A0A1I8MRE0 A0A0C9RNS9 A0A0J7MYN5 A0A1B6JEE1 A0A1B6JQV7 N6TLR8 U4UF76 A0A1A9YN98 A0A1Q3EWD5 A0A1B0C5I3 A0A1A9ZHG6 A0A2G8KPF9 A0A0K8S753 A0A0A9YB23 A0A023F1R6 A0A146LR33 A0A1B0FEL6 J9K176 A0A1S3KAT7 A0A2H8TQC5 A0A1S3IN85 A0A1A9VPB0 A0A023EZD1 A0A2S2NYZ2 H0YWL1 A0A3B3S2J6 Q91784 Q6IRM2 A0A3B3S4F8 H9G9C3 A0A2S2Q0P6 A0A2P4TAY7 A0A226N556 A0A0L7R797 F6ZGY3 F6TNW7 A0A093QU19 A0A2I0LS47 G1MS31 A0A3L8SIC2 A0A1S3A805 A0A091L5T7 V9K8S5 A0A0A0A7J7 E9PSJ3 M3W9X8 A0A384DUB0 A0A3Q7VUB6 A0A1U7RSD0 A0A091V288 W5Q1N4 A0A1S3GE59 D2H8A0 A0A1S3GE90
A0A2A4JVP0 A0A3S2LYT7 A0A194Q908 A0A212EJ78 A0A0N1INR7 A0A195FK11 A0A158P2Q9 A0A151I0F6 F4W5R8 A0A195EZL8 F4WXB4 A0A151X2K6 A0A2A3E9X3 A0A195DFE9 A0A088A2C5 A0A0N0BGY7 A0A195C3L1 E2AZU7 A0A1Y1MH00 A0A154P8G1 A0A026VV16 A0A3L8D4X3 D6WTV8 E9JC95 A0A1W4XDT4 A0A1W4XPH6 A0A1W4XE54 E0VBK4 E2BRZ7 A0A1B6E537 A0A2P8Y0R4 K7IQ94 A0A232EX98 T1PMI9 A0A1I8MRE6 A0A1I8MRG6 A0A0L7QXV6 A0A1I8MRE0 A0A0C9RNS9 A0A0J7MYN5 A0A1B6JEE1 A0A1B6JQV7 N6TLR8 U4UF76 A0A1A9YN98 A0A1Q3EWD5 A0A1B0C5I3 A0A1A9ZHG6 A0A2G8KPF9 A0A0K8S753 A0A0A9YB23 A0A023F1R6 A0A146LR33 A0A1B0FEL6 J9K176 A0A1S3KAT7 A0A2H8TQC5 A0A1S3IN85 A0A1A9VPB0 A0A023EZD1 A0A2S2NYZ2 H0YWL1 A0A3B3S2J6 Q91784 Q6IRM2 A0A3B3S4F8 H9G9C3 A0A2S2Q0P6 A0A2P4TAY7 A0A226N556 A0A0L7R797 F6ZGY3 F6TNW7 A0A093QU19 A0A2I0LS47 G1MS31 A0A3L8SIC2 A0A1S3A805 A0A091L5T7 V9K8S5 A0A0A0A7J7 E9PSJ3 M3W9X8 A0A384DUB0 A0A3Q7VUB6 A0A1U7RSD0 A0A091V288 W5Q1N4 A0A1S3GE59 D2H8A0 A0A1S3GE90
PDB
3ZFD
E-value=1.19158e-93,
Score=880
Ontologies
GO
GO:0008017
GO:0005524
GO:0003777
GO:0007018
GO:0003774
GO:0005874
GO:0019835
GO:0016021
GO:0016887
GO:0007052
GO:0005871
GO:0005815
GO:0005737
GO:0005694
GO:0005634
GO:0003677
GO:0003676
GO:0008270
GO:0000281
GO:0051256
GO:0045171
GO:0030496
GO:0005654
GO:0003690
GO:0003697
GO:0008094
GO:0006281
GO:0006259
Topology
Subcellular location
Nucleus
Chromosome
Cytoplasm
Cytoskeleton
Chromosome
Cytoplasm
Cytoskeleton
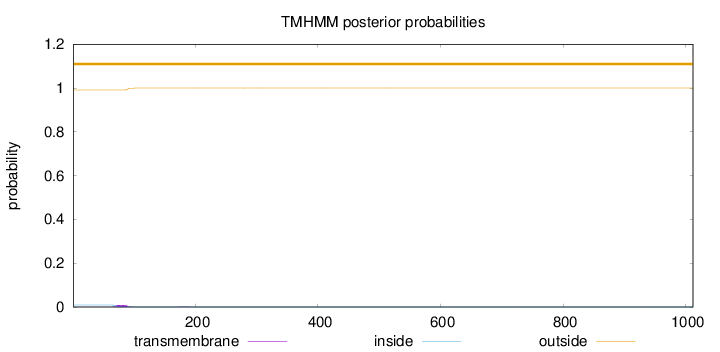
Length:
1012
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18518
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00889
outside
1 - 1012
Population Genetic Test Statistics
Pi
200.508922
Theta
21.830234
Tajima's D
-0.7452
CLR
246.843137
CSRT
0.185690715464227
Interpretation
Uncertain
