Gene
KWMTBOMO15386
Pre Gene Modal
BGIBMGA005105
Annotation
PREDICTED:_putative_epidermal_cell_surface_receptor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.197
Sequence
CDS
ATGAATTGTAAAATAGCTAACGACGAAGAAGAAGTGCCATCAACTACGTCATCACAAACACTAGAGACATTACCTCCAATCTCAACTCCTACACCAAGAATAATAGAAGTGACAGAAAACGTGACCGAAGCAGAATTGACGAAAGAAGCAGAAGTTACAACAGAAGTTCAAGAAGCTATCGAACCAAAACCACAACCTAGACTACTCAATTTAAATGTAGATGAACTTCAAAACTTCGCGAACGCACTACACAATCAGACTATCGAGAAGAGCAGTAAAAAGGGTTTGGATTTGAGTGACGTAAATCTTGATGAAGAAGACGACGAAGTTCCGAGGATACACGATCACAAGAACAAGAATATTCCGGACAAATTTTATTCAAATTTACAAGCGCCTTTCCATCATTTATTGAATAACGAACGGGGCTCGGATGAAATCGGAACGTGCAAAGAAAACGAAATCAGTTATAAGATCGGTGATCATATGGATCGTGGATGCGAAGAATCATGTGAGTGTACTCCGGGCGGCGTTTTCGAATGTTCACCGCGCTGTAAACATCCTTACATTCGCAGCGGGCGACGATTGAACGATCCCTTATGCTTTGAATCTCCTGTAGACAGCTGCTGCTCTATTATCGCTTGTTCCACTGGTAATGGAGATACTAAGCCAATGAAACTGGATGTTTGCCGCTACGGCAACGATACATACCCGGTCGGGACCAAATGGAATATCGGATGTGAACAGAGCTGCATGTGCGAACCTAATTCTGTAGCTACGTGCAAACCTCGATGCAAACAATACAGTCCGTCCGAGAAATGTATCAACGTCCAAGATCCGAATGACGCGTGCTGCGAGATCCAAGTGTGCGATGTATCACAAGACGTTCACGAAGAACCGCCTGATAACGTCACATCTTCTACGACAAGCGCTAGTGAGACGGCAAGAAGCTTTGATATGTCGGAAGAATTAAAATCGACCAGTAGACCCCTCGTCTTAGCTGAACCCATCGGCTCAGTCAAAGTACTGAACAATAACTCAGTCCAAGTCAATCTCATCCACGTGAACACAACTGAAGATCCAGTCCATTTACTATTAAGCGATGATGGCGGAAAAACATATCAAGACGTAGAACTTAAGTACTCGAATTTGATTCTTAATCTTGACGGCGGTAAAGATTACATTCTGAAAACAAAAGAAACCGGTACTAAATTTAACTTCACTATAACGACAACAGCCTTCGCTGAGAATGAAGTGAAGGTTGGAGCAGAAGATATGAGAAACGATACCAAAGAAGGGTGTTATGAAAATGGAAAGTTTTATGAAATTGGCGAAGAGTTCCATATCGGATGCTCGGAGTTGTGCGAGTGTACCGGCCCGGACAAGCGGGAGTGCGCGGCCCTGGTGTGTCCGTCGCATGTGGGCCTGGAGCTGATGTCCAAGGGCTGCGTGCGGTGGGCGCCCTCACCGCCCGCGGTGCCGCCCAACTGCTGCCCGCGGTCCGCCAGGTGCCTCAGCGATGGAACCTGCCATTACAAGGGAGTCGCTGTGCCTAACTGGAGTGAAGTCCCTCTCGACCTCACGGGCTGCGAGCAACGCTGTTTCTGCGAGAACGGAGAGCTGGACTGCCAGGAGGTCTGCTCTCCGCTACCAGCGGTTCCGCCTCAAAATTTCAGATGTCCGCCTTTGCACAGGCCGGCACCCGTCAATATATCTGACGAGGATTGTTGTAAGGAATGGGGATGTGTACCCAATGGCGAAGCACCACCGCCACCACCCGGGTTCCCTCAACAAGATTCTCGTCCCCAACAACCCTCCTTCCTTCCAACTATTCCCCCAGAGATCCACTTTCCGCAAGAGGATATTCCAGATTATGACCAGAATAACATTCATCGTCCCAGCAACCCCTTGGAGCCGGGCATACCAGGATTACCTCCAGGATTTTCAATGCCAACGAACTATGGAGAAGTTAATAAGAAATTATCTGTAATTAGTCTTCAAGCGGACTCTCCGAACAGTGTGAAGATGATGTTCGGACTTCCTCCCGTTCTGGTAGGACTACGTGGAAGTGTGGACTTGAGATATACTGATAAGGTAGACGAAGATGTATCTCAGTGGAGTTCCCAAGTGTTCGCTCCGGCTGACGAGGTGCTGACCACGCCGCGCTTGGAGTTCCGACTGACAGGCCTGAAGCCGTCGACAACGTATAAGATCCGGGCGAAGCTCTACCTCCATAACTTACCCGTCGAACCGAAAAGCGAAGTCTATACTGTTCGGACCCAGGACGCGCCGACTATCGAGACACCGGTCGAAGAGAAACGCAAAGAAATTGACAGTAAACTAACAGTTTTGGAAGTAAACGATACAACAGCACAGCTCAACTGGAGACGCTTCTCTGAGGAAGAACTGCAGTACATTGATGGCATACAAGTCAGGTATCGTCCAGTGGGCACGCCTATATACAGCATGACGGAGCTCCTCCACCACAGCAGACCTGCAGTAGAACTGCACGAGCTGCGTCCAGGAACTCCATACGAAGCCTCGCTGGTCCTGATCCCTCCTCCGCGGAGTAACACAGAACTGATAGACCCTGGTCGCGTGGAGTTTACCACTGCGCCTTATACCGACCCGTACAACTGGTCGGTGACGGTGGAGGCGTTGTCGGTGGGCGCGGGCGCGGTGCAGGTGTCGTGGCGCGGCGTGCCGGCGCCGGCCGAGCGCTGGGTGCGCGTGTACCGAGCCGCGCACTCGTGCGGGCGGCGGGCGGCGGGCGGGGCGCGGGCCGCGGGGAGGGAGCACGACGCCTTCACACTCGCCGCGAGGGACGCGCCCACCACGCTCACTCTCAGCGGCCTGGAACCGGATTCAAGATGTCGCGTTTGGCTCGAACTGTTCCTGACGAACGGGAAGGTGAAGACAAGCAACGTGCTGGAGATCAACACGAGGCCGGCGGACGAGCCTGAGGACGTGGACAATGAAATCGAAGCATCCTCAGTGTCCGGCAGCCGCAGTGCAGAGCGCGGGGACTACTACGGAGGGCTGGTCGTCGCCGGAGTGCTGGCGGCCCTCGGGGCGCTGACCTCCCTACTGCTGCTGCTCGTTGTGGTCAGACGACACCGTCCTAGATCCGTGCCCATCACACCAGTACCAACAGCTCCGAGAGAGTCGTCTCTACCTCCCTACGACAATCCTGCGTACAAACTGGAATTACAACAGGAGACAATGGATCTCTGA
Protein
MNCKIANDEEEVPSTTSSQTLETLPPISTPTPRIIEVTENVTEAELTKEAEVTTEVQEAIEPKPQPRLLNLNVDELQNFANALHNQTIEKSSKKGLDLSDVNLDEEDDEVPRIHDHKNKNIPDKFYSNLQAPFHHLLNNERGSDEIGTCKENEISYKIGDHMDRGCEESCECTPGGVFECSPRCKHPYIRSGRRLNDPLCFESPVDSCCSIIACSTGNGDTKPMKLDVCRYGNDTYPVGTKWNIGCEQSCMCEPNSVATCKPRCKQYSPSEKCINVQDPNDACCEIQVCDVSQDVHEEPPDNVTSSTTSASETARSFDMSEELKSTSRPLVLAEPIGSVKVLNNNSVQVNLIHVNTTEDPVHLLLSDDGGKTYQDVELKYSNLILNLDGGKDYILKTKETGTKFNFTITTTAFAENEVKVGAEDMRNDTKEGCYENGKFYEIGEEFHIGCSELCECTGPDKRECAALVCPSHVGLELMSKGCVRWAPSPPAVPPNCCPRSARCLSDGTCHYKGVAVPNWSEVPLDLTGCEQRCFCENGELDCQEVCSPLPAVPPQNFRCPPLHRPAPVNISDEDCCKEWGCVPNGEAPPPPPGFPQQDSRPQQPSFLPTIPPEIHFPQEDIPDYDQNNIHRPSNPLEPGIPGLPPGFSMPTNYGEVNKKLSVISLQADSPNSVKMMFGLPPVLVGLRGSVDLRYTDKVDEDVSQWSSQVFAPADEVLTTPRLEFRLTGLKPSTTYKIRAKLYLHNLPVEPKSEVYTVRTQDAPTIETPVEEKRKEIDSKLTVLEVNDTTAQLNWRRFSEEELQYIDGIQVRYRPVGTPIYSMTELLHHSRPAVELHELRPGTPYEASLVLIPPPRSNTELIDPGRVEFTTAPYTDPYNWSVTVEALSVGAGAVQVSWRGVPAPAERWVRVYRAAHSCGRRAAGGARAAGREHDAFTLAARDAPTTLTLSGLEPDSRCRVWLELFLTNGKVKTSNVLEINTRPADEPEDVDNEIEASSVSGSRSAERGDYYGGLVVAGVLAALGALTSLLLLLVVVRRHRPRSVPITPVPTAPRESSLPPYDNPAYKLELQQETMDL
Summary
Uniprot
EMBL
NWSH01000553
PCG75692.1
RSAL01000107
RVE47269.1
PCG75691.1
NEVH01019370
+ More
PNF22791.1 GL438386 EFN69059.1 LJIG01009291 KRT83374.1 PNF22793.1 KK852683 KDR18539.1 DS235751 EEB16374.1 GBHO01043784 GBHO01043780 JAF99819.1 JAF99823.1 GBHO01043779 GBHO01015135 GBHO01007720 GBHO01007711 JAF99824.1 JAG28469.1 JAG35884.1 JAG35893.1 GEBQ01031853 JAT08124.1 GEDC01005899 JAS31399.1 GBHO01043782 GBHO01015141 JAF99821.1 JAG28463.1 GBHO01015142 GBHO01015139 GBHO01015138 GBHO01007716 JAG28462.1 JAG28465.1 JAG28466.1 JAG35888.1 GEBQ01023972 JAT16005.1
PNF22791.1 GL438386 EFN69059.1 LJIG01009291 KRT83374.1 PNF22793.1 KK852683 KDR18539.1 DS235751 EEB16374.1 GBHO01043784 GBHO01043780 JAF99819.1 JAF99823.1 GBHO01043779 GBHO01015135 GBHO01007720 GBHO01007711 JAF99824.1 JAG28469.1 JAG35884.1 JAG35893.1 GEBQ01031853 JAT08124.1 GEDC01005899 JAS31399.1 GBHO01043782 GBHO01015141 JAF99821.1 JAG28463.1 GBHO01015142 GBHO01015139 GBHO01015138 GBHO01007716 JAG28462.1 JAG28465.1 JAG28466.1 JAG35888.1 GEBQ01023972 JAT16005.1
Proteomes
Pfam
PF00041 fn3
SUPFAM
SSF49265
SSF49265
Gene 3D
CDD
ProteinModelPortal
Ontologies
GO
Topology
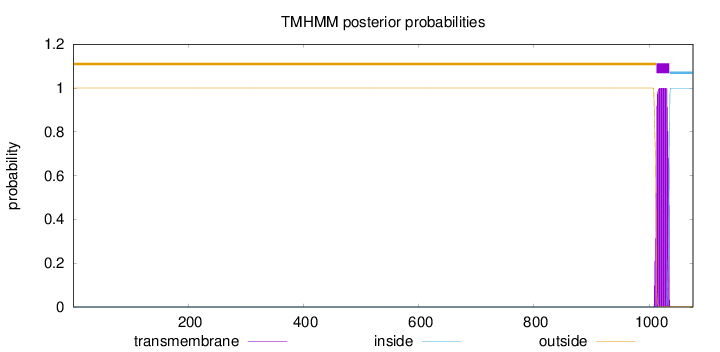
Length:
1076
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.89806
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 1012
TMhelix
1013 - 1035
inside
1036 - 1076
Population Genetic Test Statistics
Pi
211.845047
Theta
193.643349
Tajima's D
0.520879
CLR
0.734774
CSRT
0.518474076296185
Interpretation
Uncertain
