Gene
KWMTBOMO15381
Annotation
PREDICTED:_trypsin-2-like_[Bombyx_mori]
Full name
Hypodermin-A
Location in the cell
Extracellular Reliability : 1.994
Sequence
CDS
ATGTTTCCTCATTCAACTTATCTAAGTATTCCATGTGAAGGTAATTTCATGTGTGGTTCTTCAGTTTTGGGCCAAAATGTTCTTCTAACGGCGGCTCATTGTGTATGCAACTGTAATTTAAGAAAAAACAGATTAGAAGCTCTAGCTGGACATGCGTTAATCAAAAAGATGACTCTTCGACGGAGTGTTACGGCTATGACAGTTCATGCAAAGTATAATGAAAAAACAATAGAAAATGACATAGCAATATTATGGCTCGATCGTAGATTATTGTTTGGGGATAACGTGAAACGCGTTATAATTACAAAAACATTTCCATTCACAACCGAAGCCTGTATCGCTGGCTGGGGACTTATTAACGTGAGTAAAACCACTTGA
Protein
MFPHSTYLSIPCEGNFMCGSSVLGQNVLLTAAHCVCNCNLRKNRLEALAGHALIKKMTLRRSVTAMTVHAKYNEKTIENDIAILWLDRRLLFGDNVKRVIITKTFPFTTEACIAGWGLINVSKTT
Summary
Similarity
Belongs to the peptidase S1 family.
Keywords
Direct protein sequencing
Disulfide bond
Hydrolase
Protease
Secreted
Serine protease
Signal
Zymogen
Feature
propeptide Activation peptide
chain Hypodermin-A
chain Hypodermin-A
Uniprot
A0A194Q913
A0A194QDC4
A0A194RKI2
A0A212FK82
A0A212FKP7
A0A2H1V9I9
+ More
A0A2H1VHC8 A0A2A4JL27 A0A2H1WKW7 A0A212FKM8 A0A2H1W4Q5 A0A0L7LFE7 A0A2H1VR93 B0WLB8 A0A0L7KT34 A0A0T6B7T2 A0A2W1BJQ7 A0A3P9DN47 W5JDQ9 B3MWA2 A0A3Q3M8B3 A0A3S2P785 A0A0L7LG02 A0A3B5AUC8 A0A0L7KVC9 A0A1Q3FSI0 A7DYY1 A7DYX9 A7DYX7 B4Q817 Q5QBG3 A0A3S2LBD0 A0A3P8WLP4 A0A1I8Q6S0 A0A3B5AKI3 A0A3B5AW50 H2UD15 A0A3Q3LWL8 A0A3Q3M853 A0A3B5AME9 A7DYY0 H9J0R7 B4I2H7 I3KBF0 A0A3B5AMM2 A0A3B5AKR2 A0A3B4T346 A0A3P8PL82 A0A3Q3LWK4 A0A182GGY4 A0A1W7R806 Q9VXC9 Q0GSU5 A0A1L3J040 A0A1Q3FPS2 A0A226EAL5 B3NVL1 A0A067R782 B4IER6 A0A1A9W2J2 A0A0B7K512 A0A3B5AKU5 A0A1Q3FQJ5 A0A1L3J026 A0A1L3J025 A0A1L3J024 A0A1L3J041 A0A1L3J065 A0A2I0U4S9 A0A1A9W0A9 A0A1L3J049 A0A3M6U1N3 A0A1Y1KBN6 Q17IG9 H9J0R6 Q175P6 A7DYX6 A0A1W4W8M1 A7DYX5 A0A182GAC9 A0A1L9WNS2 A0A2U9C0F9 A0A1Q3FNA4 A0A1L3J027 Q5BAR4 A0A1U8QWP9 A0A1L3J028 B4R616 A0A1L3J018 A0A1L3J034 A0A3Q1GSD3 A0A3Q4GW67 A0A099ZLA0 A0A3Q1FUL4 A7DYV9 A0A1W4XLZ4 A0A0L7LF91 A0A182IT30 P35587 A0A1L3J044
A0A2H1VHC8 A0A2A4JL27 A0A2H1WKW7 A0A212FKM8 A0A2H1W4Q5 A0A0L7LFE7 A0A2H1VR93 B0WLB8 A0A0L7KT34 A0A0T6B7T2 A0A2W1BJQ7 A0A3P9DN47 W5JDQ9 B3MWA2 A0A3Q3M8B3 A0A3S2P785 A0A0L7LG02 A0A3B5AUC8 A0A0L7KVC9 A0A1Q3FSI0 A7DYY1 A7DYX9 A7DYX7 B4Q817 Q5QBG3 A0A3S2LBD0 A0A3P8WLP4 A0A1I8Q6S0 A0A3B5AKI3 A0A3B5AW50 H2UD15 A0A3Q3LWL8 A0A3Q3M853 A0A3B5AME9 A7DYY0 H9J0R7 B4I2H7 I3KBF0 A0A3B5AMM2 A0A3B5AKR2 A0A3B4T346 A0A3P8PL82 A0A3Q3LWK4 A0A182GGY4 A0A1W7R806 Q9VXC9 Q0GSU5 A0A1L3J040 A0A1Q3FPS2 A0A226EAL5 B3NVL1 A0A067R782 B4IER6 A0A1A9W2J2 A0A0B7K512 A0A3B5AKU5 A0A1Q3FQJ5 A0A1L3J026 A0A1L3J025 A0A1L3J024 A0A1L3J041 A0A1L3J065 A0A2I0U4S9 A0A1A9W0A9 A0A1L3J049 A0A3M6U1N3 A0A1Y1KBN6 Q17IG9 H9J0R6 Q175P6 A7DYX6 A0A1W4W8M1 A7DYX5 A0A182GAC9 A0A1L9WNS2 A0A2U9C0F9 A0A1Q3FNA4 A0A1L3J027 Q5BAR4 A0A1U8QWP9 A0A1L3J028 B4R616 A0A1L3J018 A0A1L3J034 A0A3Q1GSD3 A0A3Q4GW67 A0A099ZLA0 A0A3Q1FUL4 A7DYV9 A0A1W4XLZ4 A0A0L7LF91 A0A182IT30 P35587 A0A1L3J044
EC Number
3.4.21.-
Pubmed
26354079
22118469
26227816
28756777
25186727
20920257
+ More
23761445 17994087 17573377 22936249 15796745 24487278 21551351 19121390 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 16783023 27883252 24845553 30382153 28004739 17510324 28196534 16372000 19146970 7808473 7018579
23761445 17994087 17573377 22936249 15796745 24487278 21551351 19121390 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 16783023 27883252 24845553 30382153 28004739 17510324 28196534 16372000 19146970 7808473 7018579
EMBL
KQ459324
KPJ01455.1
KPJ01456.1
KQ460045
KPJ18047.1
AGBW02008116
+ More
OWR54136.1 AGBW02008010 OWR54306.1 ODYU01001393 SOQ37513.1 ODYU01002383 SOQ39812.1 NWSH01001202 PCG72143.1 ODYU01009360 SOQ53730.1 OWR54305.1 ODYU01006324 SOQ48069.1 JTDY01001347 KOB74130.1 ODYU01003944 SOQ43317.1 DS232962 DS231983 EDS27062.1 EDS30402.1 JTDY01006244 KOB66174.1 LJIG01009274 KRT83408.1 KZ150000 PZC75312.1 ADMH02001669 ETN61483.1 CH902625 EDV35247.1 RSAL01000249 RVE43502.1 KOB74131.1 JTDY01005274 KOB67168.1 GFDL01004672 JAV30373.1 AM765883 CAO78784.1 AM765881 CAO78782.1 AM765879 AM765880 CAO78780.1 CAO78781.1 CM000361 CM002910 EDX03439.1 KMY87631.1 AY752848 AAV84261.1 RSAL01000282 RVE43013.1 AM765882 CAO78783.1 BABH01010245 CH480820 EDW53972.1 AERX01014184 AERX01014185 JXUM01062824 KQ562217 KXJ76377.1 GEHC01000378 JAV47267.1 AE014298 AY118432 AAF48647.1 AAM48461.1 DQ539257 DQ539259 DQ539260 ABG02778.1 ABG02780.1 ABG02781.1 KY087128 KY087137 APG58489.1 GFDL01005572 JAV29473.1 LNIX01000005 OXA54258.1 CH954180 EDV46403.1 KK852655 KDR19292.1 CH480832 EDW46170.1 CDPU01000028 CEO52428.1 GFDL01005165 JAV29880.1 KY087112 APG58473.1 KY087106 KY087108 KY087111 KY087113 KY087117 KY087126 KY087136 KY087140 APG58472.1 KY087107 KY087139 APG58468.1 KY087134 APG58495.1 KY087143 APG58504.1 KZ506167 PKU41078.1 KY087138 APG58499.1 RCHS01002416 RMX47449.1 GEZM01091341 JAV56896.1 CH477239 EAT46500.1 BABH01010242 BABH01010243 CH477397 EAT41804.1 AM765878 CAO78779.1 AM765877 CAO78778.1 JXUM01008849 KQ560270 KXJ83415.1 KV878981 OJJ97808.1 CP026253 AWP09961.1 GFDL01006008 JAV29037.1 KY087110 KY087115 KY087127 KY087141 APG58476.1 BN001307 CBF86700.1 AACD01000039 EAA64477.1 KY087105 KY087109 KY087121 KY087125 KY087135 APG58470.1 CM000366 EDX18120.1 KY087114 APG58475.1 KY087116 APG58477.1 KL895518 KGL82641.1 AM765861 CAO78762.1 KOB74132.1 X74303 L24914 KY087133 APG58494.1
OWR54136.1 AGBW02008010 OWR54306.1 ODYU01001393 SOQ37513.1 ODYU01002383 SOQ39812.1 NWSH01001202 PCG72143.1 ODYU01009360 SOQ53730.1 OWR54305.1 ODYU01006324 SOQ48069.1 JTDY01001347 KOB74130.1 ODYU01003944 SOQ43317.1 DS232962 DS231983 EDS27062.1 EDS30402.1 JTDY01006244 KOB66174.1 LJIG01009274 KRT83408.1 KZ150000 PZC75312.1 ADMH02001669 ETN61483.1 CH902625 EDV35247.1 RSAL01000249 RVE43502.1 KOB74131.1 JTDY01005274 KOB67168.1 GFDL01004672 JAV30373.1 AM765883 CAO78784.1 AM765881 CAO78782.1 AM765879 AM765880 CAO78780.1 CAO78781.1 CM000361 CM002910 EDX03439.1 KMY87631.1 AY752848 AAV84261.1 RSAL01000282 RVE43013.1 AM765882 CAO78783.1 BABH01010245 CH480820 EDW53972.1 AERX01014184 AERX01014185 JXUM01062824 KQ562217 KXJ76377.1 GEHC01000378 JAV47267.1 AE014298 AY118432 AAF48647.1 AAM48461.1 DQ539257 DQ539259 DQ539260 ABG02778.1 ABG02780.1 ABG02781.1 KY087128 KY087137 APG58489.1 GFDL01005572 JAV29473.1 LNIX01000005 OXA54258.1 CH954180 EDV46403.1 KK852655 KDR19292.1 CH480832 EDW46170.1 CDPU01000028 CEO52428.1 GFDL01005165 JAV29880.1 KY087112 APG58473.1 KY087106 KY087108 KY087111 KY087113 KY087117 KY087126 KY087136 KY087140 APG58472.1 KY087107 KY087139 APG58468.1 KY087134 APG58495.1 KY087143 APG58504.1 KZ506167 PKU41078.1 KY087138 APG58499.1 RCHS01002416 RMX47449.1 GEZM01091341 JAV56896.1 CH477239 EAT46500.1 BABH01010242 BABH01010243 CH477397 EAT41804.1 AM765878 CAO78779.1 AM765877 CAO78778.1 JXUM01008849 KQ560270 KXJ83415.1 KV878981 OJJ97808.1 CP026253 AWP09961.1 GFDL01006008 JAV29037.1 KY087110 KY087115 KY087127 KY087141 APG58476.1 BN001307 CBF86700.1 AACD01000039 EAA64477.1 KY087105 KY087109 KY087121 KY087125 KY087135 APG58470.1 CM000366 EDX18120.1 KY087114 APG58475.1 KY087116 APG58477.1 KL895518 KGL82641.1 AM765861 CAO78762.1 KOB74132.1 X74303 L24914 KY087133 APG58494.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000218220
UP000037510
UP000002320
+ More
UP000265160 UP000000673 UP000007801 UP000261640 UP000283053 UP000261400 UP000000304 UP000265120 UP000095300 UP000005226 UP000005204 UP000001292 UP000005207 UP000261420 UP000265100 UP000069940 UP000249989 UP000000803 UP000198287 UP000008711 UP000027135 UP000091820 UP000275408 UP000008820 UP000192221 UP000184546 UP000246464 UP000000560 UP000005890 UP000257200 UP000261580 UP000053641 UP000192223 UP000075880
UP000265160 UP000000673 UP000007801 UP000261640 UP000283053 UP000261400 UP000000304 UP000265120 UP000095300 UP000005226 UP000005204 UP000001292 UP000005207 UP000261420 UP000265100 UP000069940 UP000249989 UP000000803 UP000198287 UP000008711 UP000027135 UP000091820 UP000275408 UP000008820 UP000192221 UP000184546 UP000246464 UP000000560 UP000005890 UP000257200 UP000261580 UP000053641 UP000192223 UP000075880
Interpro
IPR001254
Trypsin_dom
+ More
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR036364 SEA_dom_sf
IPR035914 Sperma_CUB_dom_sf
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR000859 CUB_dom
IPR023415 LDLR_class-A_CS
IPR000082 SEA_dom
IPR000884 TSP1_rpt
IPR036383 TSP1_rpt_sf
IPR012224 Pept_S1A_FX
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR036364 SEA_dom_sf
IPR035914 Sperma_CUB_dom_sf
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR000859 CUB_dom
IPR023415 LDLR_class-A_CS
IPR000082 SEA_dom
IPR000884 TSP1_rpt
IPR036383 TSP1_rpt_sf
IPR012224 Pept_S1A_FX
SUPFAM
ProteinModelPortal
A0A194Q913
A0A194QDC4
A0A194RKI2
A0A212FK82
A0A212FKP7
A0A2H1V9I9
+ More
A0A2H1VHC8 A0A2A4JL27 A0A2H1WKW7 A0A212FKM8 A0A2H1W4Q5 A0A0L7LFE7 A0A2H1VR93 B0WLB8 A0A0L7KT34 A0A0T6B7T2 A0A2W1BJQ7 A0A3P9DN47 W5JDQ9 B3MWA2 A0A3Q3M8B3 A0A3S2P785 A0A0L7LG02 A0A3B5AUC8 A0A0L7KVC9 A0A1Q3FSI0 A7DYY1 A7DYX9 A7DYX7 B4Q817 Q5QBG3 A0A3S2LBD0 A0A3P8WLP4 A0A1I8Q6S0 A0A3B5AKI3 A0A3B5AW50 H2UD15 A0A3Q3LWL8 A0A3Q3M853 A0A3B5AME9 A7DYY0 H9J0R7 B4I2H7 I3KBF0 A0A3B5AMM2 A0A3B5AKR2 A0A3B4T346 A0A3P8PL82 A0A3Q3LWK4 A0A182GGY4 A0A1W7R806 Q9VXC9 Q0GSU5 A0A1L3J040 A0A1Q3FPS2 A0A226EAL5 B3NVL1 A0A067R782 B4IER6 A0A1A9W2J2 A0A0B7K512 A0A3B5AKU5 A0A1Q3FQJ5 A0A1L3J026 A0A1L3J025 A0A1L3J024 A0A1L3J041 A0A1L3J065 A0A2I0U4S9 A0A1A9W0A9 A0A1L3J049 A0A3M6U1N3 A0A1Y1KBN6 Q17IG9 H9J0R6 Q175P6 A7DYX6 A0A1W4W8M1 A7DYX5 A0A182GAC9 A0A1L9WNS2 A0A2U9C0F9 A0A1Q3FNA4 A0A1L3J027 Q5BAR4 A0A1U8QWP9 A0A1L3J028 B4R616 A0A1L3J018 A0A1L3J034 A0A3Q1GSD3 A0A3Q4GW67 A0A099ZLA0 A0A3Q1FUL4 A7DYV9 A0A1W4XLZ4 A0A0L7LF91 A0A182IT30 P35587 A0A1L3J044
A0A2H1VHC8 A0A2A4JL27 A0A2H1WKW7 A0A212FKM8 A0A2H1W4Q5 A0A0L7LFE7 A0A2H1VR93 B0WLB8 A0A0L7KT34 A0A0T6B7T2 A0A2W1BJQ7 A0A3P9DN47 W5JDQ9 B3MWA2 A0A3Q3M8B3 A0A3S2P785 A0A0L7LG02 A0A3B5AUC8 A0A0L7KVC9 A0A1Q3FSI0 A7DYY1 A7DYX9 A7DYX7 B4Q817 Q5QBG3 A0A3S2LBD0 A0A3P8WLP4 A0A1I8Q6S0 A0A3B5AKI3 A0A3B5AW50 H2UD15 A0A3Q3LWL8 A0A3Q3M853 A0A3B5AME9 A7DYY0 H9J0R7 B4I2H7 I3KBF0 A0A3B5AMM2 A0A3B5AKR2 A0A3B4T346 A0A3P8PL82 A0A3Q3LWK4 A0A182GGY4 A0A1W7R806 Q9VXC9 Q0GSU5 A0A1L3J040 A0A1Q3FPS2 A0A226EAL5 B3NVL1 A0A067R782 B4IER6 A0A1A9W2J2 A0A0B7K512 A0A3B5AKU5 A0A1Q3FQJ5 A0A1L3J026 A0A1L3J025 A0A1L3J024 A0A1L3J041 A0A1L3J065 A0A2I0U4S9 A0A1A9W0A9 A0A1L3J049 A0A3M6U1N3 A0A1Y1KBN6 Q17IG9 H9J0R6 Q175P6 A7DYX6 A0A1W4W8M1 A7DYX5 A0A182GAC9 A0A1L9WNS2 A0A2U9C0F9 A0A1Q3FNA4 A0A1L3J027 Q5BAR4 A0A1U8QWP9 A0A1L3J028 B4R616 A0A1L3J018 A0A1L3J034 A0A3Q1GSD3 A0A3Q4GW67 A0A099ZLA0 A0A3Q1FUL4 A7DYV9 A0A1W4XLZ4 A0A0L7LF91 A0A182IT30 P35587 A0A1L3J044
PDB
2B9L
E-value=7.07119e-11,
Score=155
Ontologies
GO
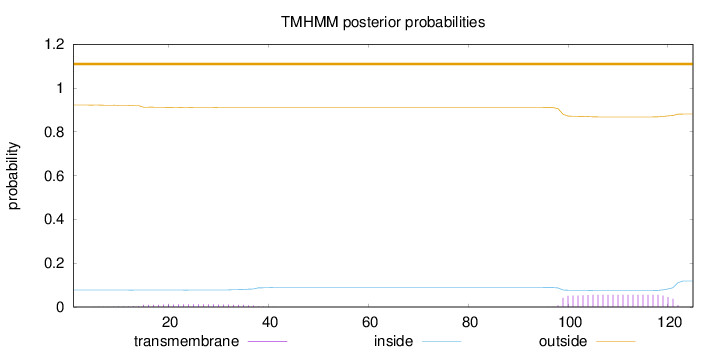
Topology
Subcellular location
Secreted
Length:
125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.50515
Exp number, first 60 AAs:
0.2659
Total prob of N-in:
0.07775
outside
1 - 125
Population Genetic Test Statistics
Pi
183.091815
Theta
161.573735
Tajima's D
1.693143
CLR
0
CSRT
0.823258837058147
Interpretation
Uncertain
