Gene
KWMTBOMO15374
Annotation
PREDICTED:_pickpocket_protein_28-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.859
Sequence
CDS
ATGGGCATCAAGCTTGTCTGTCTGCCTATCGAAGCAACTACAAATTATGGATCTCTAATTTTCAGACTTCTCTGGATTGGAGTTTTTGTGGTAATGATGAGCGCGATGTTTTACTATCTCCACCACACGTGGTTTGAGATATTGACAAAACCGCTGGTTATCAGCATGGAGTCTTCTACCTATCCCATATCAAAAATTAACTTTCCTGCTGTAGCCTTTTGTAATACGAACAGAATAAGCAGAAAGGCTTTGAAAGAATTTTCTCAGATAATGCATAGGAGCATGGCATTAAATAACGAAACAATAGAAGAAGTTGAACTTTTCTTTTTACAATATGGCCGCCTTCTAGATTATTCGTATGATGATATCTTAAGGAATCACCCCTTTTTGGATATAATAACGCTCAAAGATTTCTATTCACAAAATACATCGGAAGTCATGCAAAAGTTGGCTCCGAAATGCGACGAAATGCTACTCAGATGCGGATGGGGCAGCGAAGAAGTTAATTGCAAAACTAATTTTGACGTCCAATTGACGGTCAGGGGACATTGTTGTATATTCAACTATATTCAGGAGGACTCTGTCGACGATCCGAATTCAGCTAACAAGGACGTCAAACGGCAGTCCGTACCGGGAGCACTTTATGGATTACGTTTGGTTCTTGACCCGATGATTGATGACTACGCCTATACCGTAAACAACATACGAGGATTTGATGTTTTCCTGTTTTCACCCGGTCATTTCGCCGATTTTAGCGGGGGCAAAGTGATTTATCGCATAGTGGAGCCTGACAGAGCAGAGTTCATTGAACTATCTTCTATACAACAAAGAGCTGCTGCCGAGGTCCGGAAATATCCTGTTCGGATTAGAAAATGTTTTTTCCAAGACGAGAACGGTTCTTCACACAGGAAAACTTATACATACAACTATTGTATAATTAATTGTCGCATCAAGTCCATACAGTCGCTTTGTAAATGCACTCCTTACTTTTTGCCCGGTAAAGATAGGGCACGTACATGTACACTAGAAGATTTAAAATGCCTTAATAAATATAGGGAAAAACTTCTCTACTTGTATCCAAATGACGCCCAGGATCTTCGCGGACTTGAAACAGAAGTGGAAGATTCCCTTTTCTGTCCCAATTGCCTACCCGATTGTGAACCTATACAGCACTATGCCAAATCTTACAAGATATCTATAAATACAATTGAAGAGCACTTTGTACCATTCAGGAGTGATATGTTTGACAAAATAAACATCACCGGAAAAATCCTAGTTAGCATTTATCATTCTGCACCCAGCGGGACCTTGGATCGCCTTGATGTCGTTTCGTATTGGTTCGAAATATTAAGTAACATCGGTGGATTCTGCGGTTTTCTTTTAGGGTTTTCTCTTTTTGCCATTGTTGAAGTGTTTTACTACTTTGTCGTTCGTTTTACAATTGTTGTTACAAACGCCTACAACGTTGCTAAATAG
Protein
MGIKLVCLPIEATTNYGSLIFRLLWIGVFVVMMSAMFYYLHHTWFEILTKPLVISMESSTYPISKINFPAVAFCNTNRISRKALKEFSQIMHRSMALNNETIEEVELFFLQYGRLLDYSYDDILRNHPFLDIITLKDFYSQNTSEVMQKLAPKCDEMLLRCGWGSEEVNCKTNFDVQLTVRGHCCIFNYIQEDSVDDPNSANKDVKRQSVPGALYGLRLVLDPMIDDYAYTVNNIRGFDVFLFSPGHFADFSGGKVIYRIVEPDRAEFIELSSIQQRAAAEVRKYPVRIRKCFFQDENGSSHRKTYTYNYCIINCRIKSIQSLCKCTPYFLPGKDRARTCTLEDLKCLNKYREKLLYLYPNDAQDLRGLETEVEDSLFCPNCLPDCEPIQHYAKSYKISINTIEEHFVPFRSDMFDKINITGKILVSIYHSAPSGTLDRLDVVSYWFEILSNIGGFCGFLLGFSLFAIVEVFYYFVVRFTIVVTNAYNVAK
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
A0A2W1BV83
A0A2A4JYA3
A0A3S2LZ06
A0A212EZ61
A0A2H1VNJ6
A0A2J7PVC7
+ More
A0A195FID9 B0W5W5 A0A182YB71 A0A182R606 A0A182SN75 A0A182IT20 A0A310S8T3 A0A1S4F241 Q17HW1 A0A182HLE6 A0A182VZD6 A0A182FQ30 A0A336MKD3 Q7PXW7 A0A182L9S8 A0A139WEV4 A0A182WSJ4 A0A182NS27 A0A182Q1S7 A0A336LR92 E2C5F8 A0A182MCM8 A0A154PGC8 A0A0L7RC93 A0A1J1HVM8 A0A1I8N997 A0A026WRX6 A0A336KYA9 A0A088AUE3 W5J8I5 A0A1J1HMT7 A0A336KSU2 A0A232EPH8 A0A1A9W4F9 A0A026VZU9 F4WNX0 A0A2A3EN15 A0A310SJY6 A0A1A9YIU9 B4PV29 A0A3B0K575 A0A0L0C2G8 A0A3S2P6G5 B3LV07 E2BV58 F4WNX1 A0A336MSA4 A0A0L0BPA3 B3P6X1 B4GNR4 B3M086 A0A194QSD3 A0A1I8P9D3 A0A1I8N0Q5 A0A0L7RC42 B4HHM1 A0A1B0G526 B4QSL6 A0A1I8Q0L0 A0A1I8Q0J6 B4I015 B4QTU3 B5DYS2 A0A1W4W2L3 Q8IMV2 A0A2A4JZF1 B4LWY3 K7INU5 A0A195D4L0 B4JU76 A0A182U6B3 A0A1W4U6W3 A0A139WGL3 A0A0M5J0J4 B4NG28 D2A5I2 A0A182XCI7 A0A2J7Q831 A0A151X8F3 A0A182QKV6 B4PNC5 Q7Q1X3 A0A182UZ03 B4KIC4 Q9V9Y5 A0A232F9M1 B4K8B7 A0A2H1VX96 B4K8B6
A0A195FID9 B0W5W5 A0A182YB71 A0A182R606 A0A182SN75 A0A182IT20 A0A310S8T3 A0A1S4F241 Q17HW1 A0A182HLE6 A0A182VZD6 A0A182FQ30 A0A336MKD3 Q7PXW7 A0A182L9S8 A0A139WEV4 A0A182WSJ4 A0A182NS27 A0A182Q1S7 A0A336LR92 E2C5F8 A0A182MCM8 A0A154PGC8 A0A0L7RC93 A0A1J1HVM8 A0A1I8N997 A0A026WRX6 A0A336KYA9 A0A088AUE3 W5J8I5 A0A1J1HMT7 A0A336KSU2 A0A232EPH8 A0A1A9W4F9 A0A026VZU9 F4WNX0 A0A2A3EN15 A0A310SJY6 A0A1A9YIU9 B4PV29 A0A3B0K575 A0A0L0C2G8 A0A3S2P6G5 B3LV07 E2BV58 F4WNX1 A0A336MSA4 A0A0L0BPA3 B3P6X1 B4GNR4 B3M086 A0A194QSD3 A0A1I8P9D3 A0A1I8N0Q5 A0A0L7RC42 B4HHM1 A0A1B0G526 B4QSL6 A0A1I8Q0L0 A0A1I8Q0J6 B4I015 B4QTU3 B5DYS2 A0A1W4W2L3 Q8IMV2 A0A2A4JZF1 B4LWY3 K7INU5 A0A195D4L0 B4JU76 A0A182U6B3 A0A1W4U6W3 A0A139WGL3 A0A0M5J0J4 B4NG28 D2A5I2 A0A182XCI7 A0A2J7Q831 A0A151X8F3 A0A182QKV6 B4PNC5 Q7Q1X3 A0A182UZ03 B4KIC4 Q9V9Y5 A0A232F9M1 B4K8B7 A0A2H1VX96 B4K8B6
Pubmed
EMBL
KZ149949
PZC76700.1
NWSH01000360
PCG77001.1
RSAL01000107
RVE47278.1
+ More
AGBW02011359 OWR46786.1 ODYU01003502 SOQ42376.1 NEVH01020964 PNF20288.1 KQ981523 KYN40168.1 DS231845 EDS35952.1 KQ763832 OAD54711.1 CH477245 EAT46228.1 APCN01000896 UFQS01001084 UFQT01001084 SSX08906.1 SSX28817.1 AAAB01008987 EAA00980.5 KQ971354 KYB26357.1 AXCN02000266 UFQS01000116 UFQT01000116 SSW99925.1 SSX20305.1 GL452770 EFN76771.1 AXCM01011342 KQ434899 KZC10892.1 KQ414617 KOC68351.1 CVRI01000021 CRK91610.1 KK107119 EZA58688.1 SSX08905.1 SSX28816.1 ADMH02002100 ETN59185.1 CVRI01000009 CRK88708.1 UFQS01000961 UFQT01000961 SSX07978.1 SSX28212.1 NNAY01002936 OXU20265.1 KK107525 EZA49292.1 GL888239 EGI64128.1 KZ288205 PBC33128.1 OAD54712.1 CM000160 EDW98878.2 OUUW01000005 SPP81149.1 JRES01000984 KNC26446.1 RSAL01000289 RVE42927.1 CH902617 EDV42479.1 GL450824 EFN80350.1 EGI64129.1 SSX08907.1 SSX28818.1 JRES01001573 KNC21895.1 CH954182 EDV53791.1 CH479186 EDW38797.1 EDV42043.2 KQ461154 KPJ08433.1 KOC68350.1 CH480815 EDW43563.1 CCAG010022693 CCAG010022694 CM000364 EDX15113.1 CH480819 EDW53622.1 EDX14297.1 CM000070 EDY67632.2 AE014297 AAN14023.2 NWSH01000327 PCG77395.1 CH940650 EDW67730.2 KQ976870 KYN07823.1 CH916374 EDV91046.1 KQ971345 KYB27034.1 CP012526 ALC47831.1 CH964251 EDW83245.1 EFA05375.2 NEVH01016978 PNF24738.1 KQ982409 KYQ56656.1 AXCN02000752 EDW99207.2 AAAB01008980 EAA14549.4 CH933807 EDW13421.2 AAF57143.3 NNAY01000660 OXU27138.1 CH933806 EDW16499.2 ODYU01005000 SOQ45443.1 EDW16498.2
AGBW02011359 OWR46786.1 ODYU01003502 SOQ42376.1 NEVH01020964 PNF20288.1 KQ981523 KYN40168.1 DS231845 EDS35952.1 KQ763832 OAD54711.1 CH477245 EAT46228.1 APCN01000896 UFQS01001084 UFQT01001084 SSX08906.1 SSX28817.1 AAAB01008987 EAA00980.5 KQ971354 KYB26357.1 AXCN02000266 UFQS01000116 UFQT01000116 SSW99925.1 SSX20305.1 GL452770 EFN76771.1 AXCM01011342 KQ434899 KZC10892.1 KQ414617 KOC68351.1 CVRI01000021 CRK91610.1 KK107119 EZA58688.1 SSX08905.1 SSX28816.1 ADMH02002100 ETN59185.1 CVRI01000009 CRK88708.1 UFQS01000961 UFQT01000961 SSX07978.1 SSX28212.1 NNAY01002936 OXU20265.1 KK107525 EZA49292.1 GL888239 EGI64128.1 KZ288205 PBC33128.1 OAD54712.1 CM000160 EDW98878.2 OUUW01000005 SPP81149.1 JRES01000984 KNC26446.1 RSAL01000289 RVE42927.1 CH902617 EDV42479.1 GL450824 EFN80350.1 EGI64129.1 SSX08907.1 SSX28818.1 JRES01001573 KNC21895.1 CH954182 EDV53791.1 CH479186 EDW38797.1 EDV42043.2 KQ461154 KPJ08433.1 KOC68350.1 CH480815 EDW43563.1 CCAG010022693 CCAG010022694 CM000364 EDX15113.1 CH480819 EDW53622.1 EDX14297.1 CM000070 EDY67632.2 AE014297 AAN14023.2 NWSH01000327 PCG77395.1 CH940650 EDW67730.2 KQ976870 KYN07823.1 CH916374 EDV91046.1 KQ971345 KYB27034.1 CP012526 ALC47831.1 CH964251 EDW83245.1 EFA05375.2 NEVH01016978 PNF24738.1 KQ982409 KYQ56656.1 AXCN02000752 EDW99207.2 AAAB01008980 EAA14549.4 CH933807 EDW13421.2 AAF57143.3 NNAY01000660 OXU27138.1 CH933806 EDW16499.2 ODYU01005000 SOQ45443.1 EDW16498.2
Proteomes
UP000218220
UP000283053
UP000007151
UP000235965
UP000078541
UP000002320
+ More
UP000076408 UP000075900 UP000075901 UP000075880 UP000008820 UP000075840 UP000075920 UP000069272 UP000007062 UP000075882 UP000007266 UP000076407 UP000075884 UP000075886 UP000008237 UP000075883 UP000076502 UP000053825 UP000183832 UP000095301 UP000053097 UP000005203 UP000000673 UP000215335 UP000091820 UP000007755 UP000242457 UP000092443 UP000002282 UP000268350 UP000037069 UP000007801 UP000008711 UP000008744 UP000053240 UP000095300 UP000001292 UP000092444 UP000000304 UP000001819 UP000192221 UP000000803 UP000008792 UP000002358 UP000078542 UP000001070 UP000075902 UP000092553 UP000007798 UP000075809 UP000075903 UP000009192
UP000076408 UP000075900 UP000075901 UP000075880 UP000008820 UP000075840 UP000075920 UP000069272 UP000007062 UP000075882 UP000007266 UP000076407 UP000075884 UP000075886 UP000008237 UP000075883 UP000076502 UP000053825 UP000183832 UP000095301 UP000053097 UP000005203 UP000000673 UP000215335 UP000091820 UP000007755 UP000242457 UP000092443 UP000002282 UP000268350 UP000037069 UP000007801 UP000008711 UP000008744 UP000053240 UP000095300 UP000001292 UP000092444 UP000000304 UP000001819 UP000192221 UP000000803 UP000008792 UP000002358 UP000078542 UP000001070 UP000075902 UP000092553 UP000007798 UP000075809 UP000075903 UP000009192
Pfam
PF00858 ASC
ProteinModelPortal
A0A2W1BV83
A0A2A4JYA3
A0A3S2LZ06
A0A212EZ61
A0A2H1VNJ6
A0A2J7PVC7
+ More
A0A195FID9 B0W5W5 A0A182YB71 A0A182R606 A0A182SN75 A0A182IT20 A0A310S8T3 A0A1S4F241 Q17HW1 A0A182HLE6 A0A182VZD6 A0A182FQ30 A0A336MKD3 Q7PXW7 A0A182L9S8 A0A139WEV4 A0A182WSJ4 A0A182NS27 A0A182Q1S7 A0A336LR92 E2C5F8 A0A182MCM8 A0A154PGC8 A0A0L7RC93 A0A1J1HVM8 A0A1I8N997 A0A026WRX6 A0A336KYA9 A0A088AUE3 W5J8I5 A0A1J1HMT7 A0A336KSU2 A0A232EPH8 A0A1A9W4F9 A0A026VZU9 F4WNX0 A0A2A3EN15 A0A310SJY6 A0A1A9YIU9 B4PV29 A0A3B0K575 A0A0L0C2G8 A0A3S2P6G5 B3LV07 E2BV58 F4WNX1 A0A336MSA4 A0A0L0BPA3 B3P6X1 B4GNR4 B3M086 A0A194QSD3 A0A1I8P9D3 A0A1I8N0Q5 A0A0L7RC42 B4HHM1 A0A1B0G526 B4QSL6 A0A1I8Q0L0 A0A1I8Q0J6 B4I015 B4QTU3 B5DYS2 A0A1W4W2L3 Q8IMV2 A0A2A4JZF1 B4LWY3 K7INU5 A0A195D4L0 B4JU76 A0A182U6B3 A0A1W4U6W3 A0A139WGL3 A0A0M5J0J4 B4NG28 D2A5I2 A0A182XCI7 A0A2J7Q831 A0A151X8F3 A0A182QKV6 B4PNC5 Q7Q1X3 A0A182UZ03 B4KIC4 Q9V9Y5 A0A232F9M1 B4K8B7 A0A2H1VX96 B4K8B6
A0A195FID9 B0W5W5 A0A182YB71 A0A182R606 A0A182SN75 A0A182IT20 A0A310S8T3 A0A1S4F241 Q17HW1 A0A182HLE6 A0A182VZD6 A0A182FQ30 A0A336MKD3 Q7PXW7 A0A182L9S8 A0A139WEV4 A0A182WSJ4 A0A182NS27 A0A182Q1S7 A0A336LR92 E2C5F8 A0A182MCM8 A0A154PGC8 A0A0L7RC93 A0A1J1HVM8 A0A1I8N997 A0A026WRX6 A0A336KYA9 A0A088AUE3 W5J8I5 A0A1J1HMT7 A0A336KSU2 A0A232EPH8 A0A1A9W4F9 A0A026VZU9 F4WNX0 A0A2A3EN15 A0A310SJY6 A0A1A9YIU9 B4PV29 A0A3B0K575 A0A0L0C2G8 A0A3S2P6G5 B3LV07 E2BV58 F4WNX1 A0A336MSA4 A0A0L0BPA3 B3P6X1 B4GNR4 B3M086 A0A194QSD3 A0A1I8P9D3 A0A1I8N0Q5 A0A0L7RC42 B4HHM1 A0A1B0G526 B4QSL6 A0A1I8Q0L0 A0A1I8Q0J6 B4I015 B4QTU3 B5DYS2 A0A1W4W2L3 Q8IMV2 A0A2A4JZF1 B4LWY3 K7INU5 A0A195D4L0 B4JU76 A0A182U6B3 A0A1W4U6W3 A0A139WGL3 A0A0M5J0J4 B4NG28 D2A5I2 A0A182XCI7 A0A2J7Q831 A0A151X8F3 A0A182QKV6 B4PNC5 Q7Q1X3 A0A182UZ03 B4KIC4 Q9V9Y5 A0A232F9M1 B4K8B7 A0A2H1VX96 B4K8B6
PDB
6BQN
E-value=0.00289896,
Score=97
Ontologies
GO
PANTHER
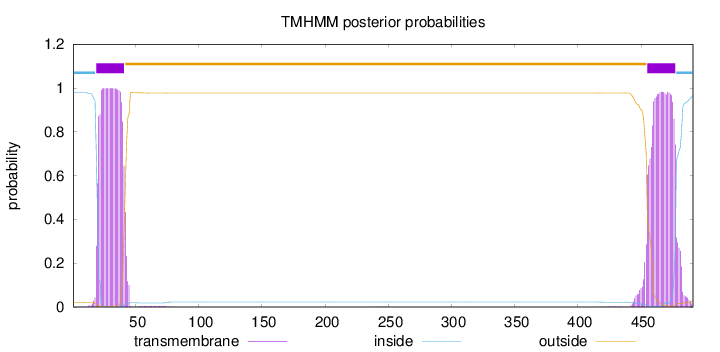
Topology
Length:
491
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.4635
Exp number, first 60 AAs:
21.98741
Total prob of N-in:
0.98019
POSSIBLE N-term signal
sequence
inside
1 - 18
TMhelix
19 - 41
outside
42 - 454
TMhelix
455 - 477
inside
478 - 491
Population Genetic Test Statistics
Pi
312.575962
Theta
165.698487
Tajima's D
2.111547
CLR
0.046326
CSRT
0.900304984750762
Interpretation
Uncertain
