Gene
KWMTBOMO15372
Annotation
reverse_transcriptase_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.369 PlasmaMembrane Reliability : 1.136
Sequence
CDS
ATGGCCCGGGCGCGTGAAGCAATCAACCTGGACGAGATTGGGGCGGAGGAAGGCCTCCGCAGCAGGCAGACGGCCAATGGGGCCAAGCTGTTAGAATGTCCCGGTGCCGATAGTCCCGGCATCGCGGAGCGTCTGGCGGCGAAGCTCCGTATGGTCTTGCCTGAGGAGGAGGTGCGGGTGCACAGGCCGGTAAAAATGGCGGAATTAAGAGTGACCGGCCTGGACGACTGCACCACCAGGGATGAGGTGAGAGCAGCCGTCGCTACCCAGGGCAACTGTGCACCGGAGAATGTGACCGTGGGTGAGCTGCGCCGTGGTTATACCGGGGCCAGCTCAGTGTGGGTGCGTTGCCCGTTGGAGACGGCTGCCTCCCTGGCTGCTCCTCCGCCCGGGGGACCGTCGGGGAATCCAGGCAGGCTGCGCGTGGGCTGGATGGCGGCCCATGTGCGCCTGCTTGAACCACGCGCCTGGCGATGCCTCCGGTGCTTTAGCACCGGACACGGTCTCGCCAGGTGCACTAGCTCGGTTGACCGCAGCGGTCTGTGTTTCCGCTGCGGCCAGCCGGGCCATAAAGCGGCGTCCTGCGCGGCCGCCCCGCACTGCGCTTTGTGCGCTGCGGCCGGGCGCGGGGCTAACCACCGAGCGGGAGGAAAGGCGTGCCTCCCGTCCGGTGATAACACCGAACGCAACCGGCACCGGAAGTCAAAGCGCCGGCAGAAGAGAAAGTTGGCCAGAGGAGCACTGGCCAACGCTGTAATCCCCGCTGCGGCGCCGGTGCCGGAGGGTGGGGGCAGCGGAAACCTGAACCACTCCGCCAGAGCTCAGGACATGCTGTCTCAGAGTATGGCGGAGTGGTCTATAGATGTGGCTGTGGTCGCCGAGCCATATTTCGTTCCCCCGGCAAATGATTGCTGGTTCGGGGACGATGGCGGCCTGGCGGCCATAGCCATAAGAAGAGACGCGGCGATCCCCCCCCTGGCGATGGTGACCAGGGGCCAAGGGTTCGTCGCGGTGCAGTTGGAAGGGACCGTCGTGATCGGGGCGTACTTCTCTCCGAACAGGCCTACTGTCGAGTTCGGGCATTTCCTGGGCGGGATCGGGGCGATTGCTCGCCGCCTCGCTCCCCGTCCAGTGATTCTCGCGGGGGATCTCAACGCGAAGTCTGTCGCATGGGGCTCCCCCCGCTCGGACGCTCGTGGTAGGCTACTGGAGAGGTGGGCGAACGCGGCCGGCCTCTGTTTGTTGAATAGAGGTTCGGTTGCGACGTGCGTGCGGTGGCAGGGCGAGTCTATTGTGGACGTTACGTTCGCGAGTCCAGCCATCGCGCGCCGTATCTGCGACTGGAAGGTTTTGGAGGGGGCGGAGACGCTGTCAGATCACCGATATATTCGGTTTGATCTCTCCGCCCGCTCCACAGTCGCAGACGTCCACGGGGACCACCCCCGTAGTGCGTCTCGGTCATTCCCCAAGTGGGCACTGAAGCGCCTGAATAAGGAGCTCCTGATGGAAGCCTCTATAGTGGCGGCGTGGGCGCCCATGCGTCCACACCCGGTCGAGGTCGAGAGCGAGGCGGGGTGGTTCCGGGACACGATGCGTCGCATTTGTGATGCGTCGATGCCCCGGATCGGCCCTCGACTTCCTAATCGCCAGGCGTACTGGTGGTCGCCCGAAATTGCGCAACTCCGCGTGGAGTGCGTTCGGGCGAGGCGCGAGTGCGCCAGACATCGCCGCCGCCGCCTGCCGCGGCGCAACGATCCGGTTGCGTTCGCGGCAGCAGAGGCCCGGCTGCACGGCGCATTTCGCGTCGCGCAGAGGGCATTGCAGCTGGCCATCAGAAGAGCCAAGAACCAACACATGGAGGGTCTCTTGGAGACGCTGGATGAGGATCCGTGGGGGCGGCCCTATCATATGGTGCGCAATAAAATGCGCCCATGGGCCCCCCCGATCACGGAGCGTCTCCAGCCTCAGCAGCTGCGGGACATCGTCTCGGCGCTGTTCCCGCAGGAGCGGGAGGGATTTGTCGCTCCCGCTATGGACGCGCCGCCGGACTACGACGGCGAAGCCCCTGCTGAGGTGCCCCGTATAACGGGGGCGGAGCTCCGTGTGGCCGTTCGCAAAATGTGCGCGAAGGACACTGCACCCGGCCCGGATGGTGTCCATGGCCGGGTTTGGGCCTTGGCTCTTGGTGCCCTAGGGGACCGACTCTTGAGACTTTACAACTCCTGCCTCGAGTCGGGACGGTTTCCTTCATCGTGGAGGACGGGCAGACTCGTGCTGTTGAGAAAGGAGGGGCGCCCGGCGGATACTGCCGCCGGGTACCGTCCCATCGTGTTGCTGGATGAGGTGGGCAAGCTGCTGGAACGCATTCTGGCAGCCCGCATCATCCAGCATCTAGTCGGGGTGGGACCCGATCTGTCGGCGGAGCAGTATGGCTTCCGAGAGGGCCGCTCAACGGTAGACGCGATCCTTCGCGTGCGGGCCCTCTCGGAGGAGGCCGTCTCCAGGGGTGGGGTGGCTTTGGCGGTGTCGCTCGATATCGCCAATGCGTTTAACACCCTGCCCTGGTCCGTGATAGGGGGGGCGCTGGAACGACATAGAGTGCCCCCCTACCTTCGCCGGCTGGTGGGTTCCTACTTAGAGGACAGATCGGTCACGTGTACCGGACACGGTGGGATCCTGCACCGGTTCCCGGTCGTGCGCGGTGTTCCACAGGGGTCGGTGCTCGGCCCCCTTTTGTGGAATATCGGGTATGACTGGGTGCTGAGAGGTGCCCTCCTCCCGGGCCTGAGCGTTATCTGTTACGCAGACGACACGTTGGTCGTGGCCCGGGGGGGAGTTTTGCTGAGTCTGCCCGTCTTGCTACGGCTGGGGCCCCGGAGAGTGCCACCTGTCGATGCCCATATCGTGGTTGGAGGCGTCCATATCGGGGTCGGGGTGCAGTTGAAGTACCTCGGCCTCATTCTGGACAGTCGTTGGACCTTCCGTGCTCACTTTCAGAACCTGGTCCCTCGTTTGTTGGGGGTGGCCGGCGCGTTAAGCCGGCTTCTGCCCAACGTCGGGGGGCCTGACCAGGTGACGCGCCGTCTCTATACGGGGGTGGTGCGATCAATGGCCCTATACGGGGCGCCCGTGTGGGGCCAGTCCCTGGCCGTGGGGGTGGCGAAGCTGCTGCAACGGCCGCAACGCACCATCGCGGTCAGGGTCATCCGTGGTTATCGCACCATCTCCTTTGAGGCGGCGTGTGTACTGGCTGGGACGCCGCCTTGGGTTCTGGAAGCGGAGGCGCTCGCCGCTGACTATAAGTGGCGGGCTGACCTTCGTGCCCGGGGCGTGGCGCGTCCCAGCCCCAGTGTGGTCAGAGCGCGGAGGGGCCAATCTCGGCGGTCCGTGCTGGAGTCATGGTCCAGACGGCTGGCCGATCCTTCGGCTGGTCGTAGGACCGTCGAGGCGATTCGCCCGGTCCTTGTGGATTGGGTGAATCGTGACAGAGGACGCCTCACTTTCCGGCTCACGCAGGTGCTCACTGGGCATGGTTGTTTCGGTGAGTTCCTGCACCGGATCGGAGCCGAGCCGACGGCAGAGTGCCACCATTGTGGTTGCGACTTGGACACGGCAGAGCATACGCTCGTCGCCTGCCCCGCATGGGAGGGGTGGCGCCGTGTCCTCGTCGCAAAAATAGGAACCGACTTGTCGTTGCCGAGTGTTGTGGCATCGATGCTCGGCGACGACGAGTCGTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGCGTGAGAGACGCACAATCCCGCCGCCGTCGAGCGGGGGCCAGGGAGGCGGATCTCGCCCAAGCCCTGGCCCTCTAA
Protein
MARAREAINLDEIGAEEGLRSRQTANGAKLLECPGADSPGIAERLAAKLRMVLPEEEVRVHRPVKMAELRVTGLDDCTTRDEVRAAVATQGNCAPENVTVGELRRGYTGASSVWVRCPLETAASLAAPPPGGPSGNPGRLRVGWMAAHVRLLEPRAWRCLRCFSTGHGLARCTSSVDRSGLCFRCGQPGHKAASCAAAPHCALCAAAGRGANHRAGGKACLPSGDNTERNRHRKSKRRQKRKLARGALANAVIPAAAPVPEGGGSGNLNHSARAQDMLSQSMAEWSIDVAVVAEPYFVPPANDCWFGDDGGLAAIAIRRDAAIPPLAMVTRGQGFVAVQLEGTVVIGAYFSPNRPTVEFGHFLGGIGAIARRLAPRPVILAGDLNAKSVAWGSPRSDARGRLLERWANAAGLCLLNRGSVATCVRWQGESIVDVTFASPAIARRICDWKVLEGAETLSDHRYIRFDLSARSTVADVHGDHPRSASRSFPKWALKRLNKELLMEASIVAAWAPMRPHPVEVESEAGWFRDTMRRICDASMPRIGPRLPNRQAYWWSPEIAQLRVECVRARRECARHRRRRLPRRNDPVAFAAAEARLHGAFRVAQRALQLAIRRAKNQHMEGLLETLDEDPWGRPYHMVRNKMRPWAPPITERLQPQQLRDIVSALFPQEREGFVAPAMDAPPDYDGEAPAEVPRITGAELRVAVRKMCAKDTAPGPDGVHGRVWALALGALGDRLLRLYNSCLESGRFPSSWRTGRLVLLRKEGRPADTAAGYRPIVLLDEVGKLLERILAARIIQHLVGVGPDLSAEQYGFREGRSTVDAILRVRALSEEAVSRGGVALAVSLDIANAFNTLPWSVIGGALERHRVPPYLRRLVGSYLEDRSVTCTGHGGILHRFPVVRGVPQGSVLGPLLWNIGYDWVLRGALLPGLSVICYADDTLVVARGGVLLSLPVLLRLGPRRVPPVDAHIVVGGVHIGVGVQLKYLGLILDSRWTFRAHFQNLVPRLLGVAGALSRLLPNVGGPDQVTRRLYTGVVRSMALYGAPVWGQSLAVGVAKLLQRPQRTIAVRVIRGYRTISFEAACVLAGTPPWVLEAEALAADYKWRADLRARGVARPSPSVVRARRGQSRRSVLESWSRRLADPSAGRRTVEAIRPVLVDWVNRDRGRLTFRLTQVLTGHGCFGEFLHRIGAEPTAECHHCGCDLDTAEHTLVACPAWEGWRRVLVAKIGTDLSLPSVVASMLGDDESWKAMLDFCECTISQKEAAGRVRDAQSRRRRAGAREADLAQALAL
Summary
Uniprot
Q868Q4
A0A2W1C3J5
A0A0J7KIY5
A0A0J7KYD2
A0A0J7MY17
A0A2S2PEZ7
+ More
X1WJY4 J9LVU7 A0A0J7KQY4 J9JWJ3 A0A0J7KIS5 A0A2H8TIE5 T1DCM0 A0A2S2NQQ6 A0A2M4CS98 A0A2M4ADW9 J9KJG8 J9KNN8 A0A2M4BC18 A0A2M4BC24 W8ADE4 W8ANK7 A0A034WR48 A0A142LX45 J9KT61 J9KUM6 A0A2M4CJ33 Q868S8 J9LQQ1 J9JY94 Q868S0 A0A2M4BBA7 Q868S2
X1WJY4 J9LVU7 A0A0J7KQY4 J9JWJ3 A0A0J7KIS5 A0A2H8TIE5 T1DCM0 A0A2S2NQQ6 A0A2M4CS98 A0A2M4ADW9 J9KJG8 J9KNN8 A0A2M4BC18 A0A2M4BC24 W8ADE4 W8ANK7 A0A034WR48 A0A142LX45 J9KT61 J9KUM6 A0A2M4CJ33 Q868S8 J9LQQ1 J9JY94 Q868S0 A0A2M4BBA7 Q868S2
EMBL
AB090825
BAC57926.1
KZ149896
PZC78733.1
LBMM01006954
KMQ90166.1
+ More
LBMM01002052 KMQ95336.1 LBMM01014122 KMQ85315.1 GGMR01015159 MBY27778.1 ABLF02011238 ABLF02034925 LBMM01004262 KMQ92649.1 ABLF02013358 ABLF02013361 ABLF02054869 LBMM01006977 KMQ90149.1 GFXV01002090 MBW13895.1 GALA01001777 JAA93075.1 GGMR01006838 MBY19457.1 GGFL01004044 MBW68222.1 GGFK01005660 MBW38981.1 ABLF02041886 ABLF02023257 GGFJ01001453 MBW50594.1 GGFJ01001454 MBW50595.1 GAMC01020519 GAMC01020515 JAB86040.1 GAMC01020517 JAB86038.1 GAKP01002292 JAC56660.1 KU543683 AMS38371.1 ABLF02009073 ABLF02030702 ABLF02042963 ABLF02041557 GGFL01001192 MBW65370.1 AB090813 BAC57902.1 ABLF02030663 ABLF02041312 ABLF02007989 AB090817 BAC57910.1 GGFJ01001194 MBW50335.1 AB090816 BAC57908.1
LBMM01002052 KMQ95336.1 LBMM01014122 KMQ85315.1 GGMR01015159 MBY27778.1 ABLF02011238 ABLF02034925 LBMM01004262 KMQ92649.1 ABLF02013358 ABLF02013361 ABLF02054869 LBMM01006977 KMQ90149.1 GFXV01002090 MBW13895.1 GALA01001777 JAA93075.1 GGMR01006838 MBY19457.1 GGFL01004044 MBW68222.1 GGFK01005660 MBW38981.1 ABLF02041886 ABLF02023257 GGFJ01001453 MBW50594.1 GGFJ01001454 MBW50595.1 GAMC01020519 GAMC01020515 JAB86040.1 GAMC01020517 JAB86038.1 GAKP01002292 JAC56660.1 KU543683 AMS38371.1 ABLF02009073 ABLF02030702 ABLF02042963 ABLF02041557 GGFL01001192 MBW65370.1 AB090813 BAC57902.1 ABLF02030663 ABLF02041312 ABLF02007989 AB090817 BAC57910.1 GGFJ01001194 MBW50335.1 AB090816 BAC57908.1
Proteomes
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
Q868Q4
A0A2W1C3J5
A0A0J7KIY5
A0A0J7KYD2
A0A0J7MY17
A0A2S2PEZ7
+ More
X1WJY4 J9LVU7 A0A0J7KQY4 J9JWJ3 A0A0J7KIS5 A0A2H8TIE5 T1DCM0 A0A2S2NQQ6 A0A2M4CS98 A0A2M4ADW9 J9KJG8 J9KNN8 A0A2M4BC18 A0A2M4BC24 W8ADE4 W8ANK7 A0A034WR48 A0A142LX45 J9KT61 J9KUM6 A0A2M4CJ33 Q868S8 J9LQQ1 J9JY94 Q868S0 A0A2M4BBA7 Q868S2
X1WJY4 J9LVU7 A0A0J7KQY4 J9JWJ3 A0A0J7KIS5 A0A2H8TIE5 T1DCM0 A0A2S2NQQ6 A0A2M4CS98 A0A2M4ADW9 J9KJG8 J9KNN8 A0A2M4BC18 A0A2M4BC24 W8ADE4 W8ANK7 A0A034WR48 A0A142LX45 J9KT61 J9KUM6 A0A2M4CJ33 Q868S8 J9LQQ1 J9JY94 Q868S0 A0A2M4BBA7 Q868S2
PDB
1WDU
E-value=1.62545e-10,
Score=164
Ontologies
GO
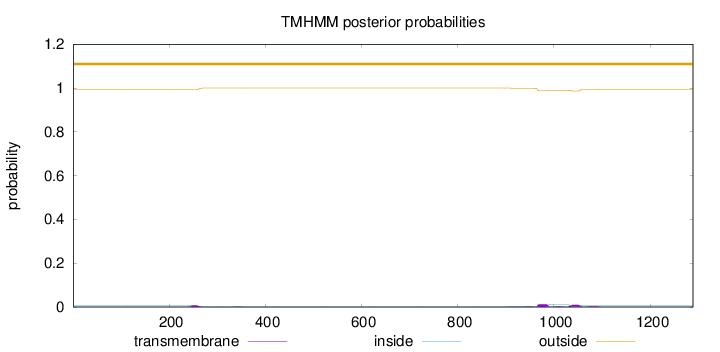
Topology
Length:
1289
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.770049999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00743
outside
1 - 1289
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
76.493494
CSRT
0
Interpretation
Uncertain
