Gene
KWMTBOMO15369
Pre Gene Modal
BGIBMGA005110
Annotation
PREDICTED:_protein_UXT_homolog_[Amyelois_transitella]
Full name
Mitochondrial inner membrane protease subunit
Location in the cell
Nuclear Reliability : 2.764
Sequence
CDS
ATGGCACAAACAAATATAGACGACACCGTAACAAGATACGAAAATTTCATAAACGAAGTACTAAAAGAAGACCTCAGAGAACTAGAAACGAAATTAGAGTTTTTTAATAATGAAGTTTCAGATTTAATACAACAGAAGCACACATTAAAAGTGTTAACAAACAAAAATATTCATCCCACCGGTTTCAAGACACAAGTTAACGTTGGTTGTAATTTCTTCATGGAGGCGTCAGTGTCCAAAACCGATACGTTGCTTGTGAACATCGGATTGAATCACTACATAGAATTTAAATTATCAGAAGCCAACAGATTTTTGGAGGCGAGAATAAAGTCTTATGAAAAAAAATCTCATGAGGTACGTCTCAGGGCAGCACAGGTTTCTGCACACATTAAGTTGATGTTATACGGTATTTCCGAATTACAAAACAGAAACAATAGTGATGGTTCTAGTACTTAA
Protein
MAQTNIDDTVTRYENFINEVLKEDLRELETKLEFFNNEVSDLIQQKHTLKVLTNKNIHPTGFKTQVNVGCNFFMEASVSKTDTLLVNIGLNHYIEFKLSEANRFLEARIKSYEKKSHEVRLRAAQVSAHIKLMLYGISELQNRNNSDGSST
Summary
Similarity
Belongs to the peptidase S1 family.
Belongs to the peptidase S26 family.
Belongs to the peptidase S26 family.
Uniprot
H9J6G8
A0A2W1BI71
A0A2A4JSC7
A0A212FK84
A0A1E1WRP8
S4PFQ4
+ More
A0A2H1VPX9 A0A0L7KXV2 A0A3S2L378 A0A194RL34 A0A194Q7E9 A0A0L7QTX5 A0A088ATL0 A0A2A3EHZ8 A0A0N0BBG0 A0A154P4D9 A0A310SQ89 A0A026WVC2 A0A067QXX1 B3N5Q2 A0A195CG93 A0A0J9R3K2 B4HXN3 B4LV42 Q9VJP9 T1H9Y4 A0A2J7QZ13 A0A1W4VKP1 E2AG35 B4NYD0 A0A1B6J4Y7 K7JKY3 A0A023F9T7 A0A1B6LDL4 A0A0J7KFL9 E9ILP8 A0A0C9QWP7 A0A151XF60 A0A195DTJ5 B3MK60 E0W063 Q29KT7 F4WF82 A0A2M4AZ04 B4GSU2 A0A182ME64 A0A2P8YGC5 E2C992 A0A158NP47 T1DRW3 B4KLG1 A0A182RWJ8 A0A2M4AYU6 B4JQS7 A0A2S2Q8I3 A0A195F9J3 A0A3B0JHY8 A0A1B6CV90 A0A195AYS7 A0A2M4C2R3 A0A0M4ENR7 A0A182JBI8 A0A2M4C2Y4 A0A1B6F8T0 A0A2M4C3N5 A0A182T1W0 A0A182FA18 A0A2M4C2W6 A0A2M3ZDE7 B4MZ94 A0A2M4C2R5 U5ES69 A0A293LAA3 B0WAF8 A0A1B0G1N6 A0A1Q3FD04 A0A1A9VA21 A0A1A9ZLY9 A0A1S8VZ34 A0A0A9Y9X3 A0A336LXH0 A0A2R7X1K6 A0A182NG38 A0A084VTG7 H3A8W3 A0A3B1IER3 A0A1A9W7W2 W5JBC4 A0A1S8VYD1 A0A1A9XHL7 A0A1B0B1N6 Q7PSN0 A0A023EGT7 A0A1S3CWH2 A0A182HA06 W5UC07 A0A182TJK3 A0A3F2YSU2 A0A1L8DUU4 A0A1Y9IZX9
A0A2H1VPX9 A0A0L7KXV2 A0A3S2L378 A0A194RL34 A0A194Q7E9 A0A0L7QTX5 A0A088ATL0 A0A2A3EHZ8 A0A0N0BBG0 A0A154P4D9 A0A310SQ89 A0A026WVC2 A0A067QXX1 B3N5Q2 A0A195CG93 A0A0J9R3K2 B4HXN3 B4LV42 Q9VJP9 T1H9Y4 A0A2J7QZ13 A0A1W4VKP1 E2AG35 B4NYD0 A0A1B6J4Y7 K7JKY3 A0A023F9T7 A0A1B6LDL4 A0A0J7KFL9 E9ILP8 A0A0C9QWP7 A0A151XF60 A0A195DTJ5 B3MK60 E0W063 Q29KT7 F4WF82 A0A2M4AZ04 B4GSU2 A0A182ME64 A0A2P8YGC5 E2C992 A0A158NP47 T1DRW3 B4KLG1 A0A182RWJ8 A0A2M4AYU6 B4JQS7 A0A2S2Q8I3 A0A195F9J3 A0A3B0JHY8 A0A1B6CV90 A0A195AYS7 A0A2M4C2R3 A0A0M4ENR7 A0A182JBI8 A0A2M4C2Y4 A0A1B6F8T0 A0A2M4C3N5 A0A182T1W0 A0A182FA18 A0A2M4C2W6 A0A2M3ZDE7 B4MZ94 A0A2M4C2R5 U5ES69 A0A293LAA3 B0WAF8 A0A1B0G1N6 A0A1Q3FD04 A0A1A9VA21 A0A1A9ZLY9 A0A1S8VZ34 A0A0A9Y9X3 A0A336LXH0 A0A2R7X1K6 A0A182NG38 A0A084VTG7 H3A8W3 A0A3B1IER3 A0A1A9W7W2 W5JBC4 A0A1S8VYD1 A0A1A9XHL7 A0A1B0B1N6 Q7PSN0 A0A023EGT7 A0A1S3CWH2 A0A182HA06 W5UC07 A0A182TJK3 A0A3F2YSU2 A0A1L8DUU4 A0A1Y9IZX9
EC Number
3.4.21.-
Pubmed
19121390
28756777
22118469
23622113
26227816
26354079
+ More
24508170 30249741 24845553 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17550304 20075255 25474469 21282665 20566863 15632085 21719571 29403074 21347285 25401762 26823975 24438588 9215903 25329095 20920257 23761445 12364791 14747013 17210077 24945155 26483478 23127152
24508170 30249741 24845553 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17550304 20075255 25474469 21282665 20566863 15632085 21719571 29403074 21347285 25401762 26823975 24438588 9215903 25329095 20920257 23761445 12364791 14747013 17210077 24945155 26483478 23127152
EMBL
BABH01019700
KZ150186
PZC72459.1
PZC72460.1
NWSH01000771
PCG74310.1
+ More
AGBW02008116 OWR54142.1 GDQN01001402 JAT89652.1 GAIX01006470 JAA86090.1 ODYU01003727 SOQ42870.1 JTDY01004589 KOB67980.1 RSAL01000193 RVE44616.1 KQ460045 KPJ18035.1 KQ459324 KPJ01467.1 KQ414740 KOC62009.1 KZ288239 PBC31328.1 KQ436043 KOX67484.1 KQ434809 KZC06713.1 KQ761076 OAD58238.1 KK107087 QOIP01000001 EZA59918.1 RLU26633.1 KK852844 KDR15140.1 CH954177 EDV58011.1 KQ977791 KYM99759.1 CM002910 KMY90354.1 CH480818 EDW51813.1 CH940649 EDW64302.1 AE014134 BT023678 KX531617 AAF53446.3 AAY85078.1 AGB92999.1 ANY27427.1 ACPB03022067 NEVH01009080 PNF33826.1 GL439266 EFN67526.1 CM000157 EDW89766.1 GECU01013474 JAS94232.1 GBBI01000456 JAC18256.1 GEBQ01018195 JAT21782.1 LBMM01008255 KMQ89021.1 GL764129 EFZ18267.1 GBYB01005097 GBYB01006004 JAG74864.1 JAG75771.1 KQ982215 KYQ58908.1 KQ980419 KYN16107.1 CH902620 EDV31478.2 DS235857 EEB19019.1 CH379061 EAL33087.2 GL888115 EGI67133.1 GGFK01012712 MBW46033.1 CH479189 EDW25451.1 AXCM01003900 PYGN01000615 PSN43305.1 GL453802 EFN75499.1 ADTU01022161 GAMD01000470 JAB01121.1 CH933807 EDW12842.1 GGFK01012655 MBW45976.1 CH916372 EDV99257.1 GGMS01004319 MBY73522.1 KQ981727 KYN36882.1 OUUW01000006 SPP81815.1 GEDC01019898 GEDC01002134 JAS17400.1 JAS35164.1 KQ976703 KYM77192.1 GGFJ01010476 MBW59617.1 CP012523 ALC38107.1 GGFJ01010478 MBW59619.1 GECZ01023132 JAS46637.1 GGFJ01010477 MBW59618.1 GGFJ01010474 MBW59615.1 GGFM01005768 MBW26519.1 CH963913 EDW77367.1 GGFJ01010475 MBW59616.1 GANO01003384 JAB56487.1 GFWV01000116 MAA24846.1 DS231871 EDS41194.1 CCAG010010949 GFDL01009608 JAV25437.1 LYON01000418 OON07329.1 GBHO01015711 GBRD01011370 GDHC01015667 JAG27893.1 JAG54454.1 JAQ02962.1 UFQS01000258 UFQT01000258 SSX02123.1 SSX22500.1 KK856407 PTY25693.1 ATLV01016353 KE525079 KFB41261.1 AFYH01231288 AFYH01231289 ADMH02001966 ETN60155.1 OON07328.1 JXJN01007216 AAAB01008817 EAA05334.5 GAPW01005553 JAC08045.1 JXUM01121971 KQ566556 KXJ70034.1 JT410561 AHH39114.1 APCN01002634 GFDF01004029 JAV10055.1
AGBW02008116 OWR54142.1 GDQN01001402 JAT89652.1 GAIX01006470 JAA86090.1 ODYU01003727 SOQ42870.1 JTDY01004589 KOB67980.1 RSAL01000193 RVE44616.1 KQ460045 KPJ18035.1 KQ459324 KPJ01467.1 KQ414740 KOC62009.1 KZ288239 PBC31328.1 KQ436043 KOX67484.1 KQ434809 KZC06713.1 KQ761076 OAD58238.1 KK107087 QOIP01000001 EZA59918.1 RLU26633.1 KK852844 KDR15140.1 CH954177 EDV58011.1 KQ977791 KYM99759.1 CM002910 KMY90354.1 CH480818 EDW51813.1 CH940649 EDW64302.1 AE014134 BT023678 KX531617 AAF53446.3 AAY85078.1 AGB92999.1 ANY27427.1 ACPB03022067 NEVH01009080 PNF33826.1 GL439266 EFN67526.1 CM000157 EDW89766.1 GECU01013474 JAS94232.1 GBBI01000456 JAC18256.1 GEBQ01018195 JAT21782.1 LBMM01008255 KMQ89021.1 GL764129 EFZ18267.1 GBYB01005097 GBYB01006004 JAG74864.1 JAG75771.1 KQ982215 KYQ58908.1 KQ980419 KYN16107.1 CH902620 EDV31478.2 DS235857 EEB19019.1 CH379061 EAL33087.2 GL888115 EGI67133.1 GGFK01012712 MBW46033.1 CH479189 EDW25451.1 AXCM01003900 PYGN01000615 PSN43305.1 GL453802 EFN75499.1 ADTU01022161 GAMD01000470 JAB01121.1 CH933807 EDW12842.1 GGFK01012655 MBW45976.1 CH916372 EDV99257.1 GGMS01004319 MBY73522.1 KQ981727 KYN36882.1 OUUW01000006 SPP81815.1 GEDC01019898 GEDC01002134 JAS17400.1 JAS35164.1 KQ976703 KYM77192.1 GGFJ01010476 MBW59617.1 CP012523 ALC38107.1 GGFJ01010478 MBW59619.1 GECZ01023132 JAS46637.1 GGFJ01010477 MBW59618.1 GGFJ01010474 MBW59615.1 GGFM01005768 MBW26519.1 CH963913 EDW77367.1 GGFJ01010475 MBW59616.1 GANO01003384 JAB56487.1 GFWV01000116 MAA24846.1 DS231871 EDS41194.1 CCAG010010949 GFDL01009608 JAV25437.1 LYON01000418 OON07329.1 GBHO01015711 GBRD01011370 GDHC01015667 JAG27893.1 JAG54454.1 JAQ02962.1 UFQS01000258 UFQT01000258 SSX02123.1 SSX22500.1 KK856407 PTY25693.1 ATLV01016353 KE525079 KFB41261.1 AFYH01231288 AFYH01231289 ADMH02001966 ETN60155.1 OON07328.1 JXJN01007216 AAAB01008817 EAA05334.5 GAPW01005553 JAC08045.1 JXUM01121971 KQ566556 KXJ70034.1 JT410561 AHH39114.1 APCN01002634 GFDF01004029 JAV10055.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000283053
UP000053240
+ More
UP000053268 UP000053825 UP000005203 UP000242457 UP000053105 UP000076502 UP000053097 UP000279307 UP000027135 UP000008711 UP000078542 UP000001292 UP000008792 UP000000803 UP000015103 UP000235965 UP000192221 UP000000311 UP000002282 UP000002358 UP000036403 UP000075809 UP000078492 UP000007801 UP000009046 UP000001819 UP000007755 UP000008744 UP000075883 UP000245037 UP000008237 UP000005205 UP000009192 UP000075900 UP000001070 UP000078541 UP000268350 UP000078540 UP000092553 UP000075880 UP000075901 UP000069272 UP000007798 UP000002320 UP000092444 UP000078200 UP000092445 UP000189906 UP000075884 UP000030765 UP000008672 UP000018467 UP000091820 UP000000673 UP000092443 UP000092460 UP000007062 UP000079169 UP000069940 UP000249989 UP000221080 UP000075902 UP000075840 UP000076407
UP000053268 UP000053825 UP000005203 UP000242457 UP000053105 UP000076502 UP000053097 UP000279307 UP000027135 UP000008711 UP000078542 UP000001292 UP000008792 UP000000803 UP000015103 UP000235965 UP000192221 UP000000311 UP000002282 UP000002358 UP000036403 UP000075809 UP000078492 UP000007801 UP000009046 UP000001819 UP000007755 UP000008744 UP000075883 UP000245037 UP000008237 UP000005205 UP000009192 UP000075900 UP000001070 UP000078541 UP000268350 UP000078540 UP000092553 UP000075880 UP000075901 UP000069272 UP000007798 UP000002320 UP000092444 UP000078200 UP000092445 UP000189906 UP000075884 UP000030765 UP000008672 UP000018467 UP000091820 UP000000673 UP000092443 UP000092460 UP000007062 UP000079169 UP000069940 UP000249989 UP000221080 UP000075902 UP000075840 UP000076407
PRIDE
Interpro
IPR011599
PFD_alpha_archaea
+ More
IPR009053 Prefoldin
IPR003994 UXT
IPR004127 Prefoldin_subunit_alpha
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR033116 TRYPSIN_SER
IPR039732 Hub1/Ubl5
IPR029071 Ubiquitin-like_domsf
IPR015927 Peptidase_S24_S26A/B/C
IPR019758 Pept_S26A_signal_pept_1_CS
IPR036286 LexA/Signal_pep-like_sf
IPR000223 Pept_S26A_signal_pept_1
IPR009053 Prefoldin
IPR003994 UXT
IPR004127 Prefoldin_subunit_alpha
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR033116 TRYPSIN_SER
IPR039732 Hub1/Ubl5
IPR029071 Ubiquitin-like_domsf
IPR015927 Peptidase_S24_S26A/B/C
IPR019758 Pept_S26A_signal_pept_1_CS
IPR036286 LexA/Signal_pep-like_sf
IPR000223 Pept_S26A_signal_pept_1
Gene 3D
CDD
ProteinModelPortal
H9J6G8
A0A2W1BI71
A0A2A4JSC7
A0A212FK84
A0A1E1WRP8
S4PFQ4
+ More
A0A2H1VPX9 A0A0L7KXV2 A0A3S2L378 A0A194RL34 A0A194Q7E9 A0A0L7QTX5 A0A088ATL0 A0A2A3EHZ8 A0A0N0BBG0 A0A154P4D9 A0A310SQ89 A0A026WVC2 A0A067QXX1 B3N5Q2 A0A195CG93 A0A0J9R3K2 B4HXN3 B4LV42 Q9VJP9 T1H9Y4 A0A2J7QZ13 A0A1W4VKP1 E2AG35 B4NYD0 A0A1B6J4Y7 K7JKY3 A0A023F9T7 A0A1B6LDL4 A0A0J7KFL9 E9ILP8 A0A0C9QWP7 A0A151XF60 A0A195DTJ5 B3MK60 E0W063 Q29KT7 F4WF82 A0A2M4AZ04 B4GSU2 A0A182ME64 A0A2P8YGC5 E2C992 A0A158NP47 T1DRW3 B4KLG1 A0A182RWJ8 A0A2M4AYU6 B4JQS7 A0A2S2Q8I3 A0A195F9J3 A0A3B0JHY8 A0A1B6CV90 A0A195AYS7 A0A2M4C2R3 A0A0M4ENR7 A0A182JBI8 A0A2M4C2Y4 A0A1B6F8T0 A0A2M4C3N5 A0A182T1W0 A0A182FA18 A0A2M4C2W6 A0A2M3ZDE7 B4MZ94 A0A2M4C2R5 U5ES69 A0A293LAA3 B0WAF8 A0A1B0G1N6 A0A1Q3FD04 A0A1A9VA21 A0A1A9ZLY9 A0A1S8VZ34 A0A0A9Y9X3 A0A336LXH0 A0A2R7X1K6 A0A182NG38 A0A084VTG7 H3A8W3 A0A3B1IER3 A0A1A9W7W2 W5JBC4 A0A1S8VYD1 A0A1A9XHL7 A0A1B0B1N6 Q7PSN0 A0A023EGT7 A0A1S3CWH2 A0A182HA06 W5UC07 A0A182TJK3 A0A3F2YSU2 A0A1L8DUU4 A0A1Y9IZX9
A0A2H1VPX9 A0A0L7KXV2 A0A3S2L378 A0A194RL34 A0A194Q7E9 A0A0L7QTX5 A0A088ATL0 A0A2A3EHZ8 A0A0N0BBG0 A0A154P4D9 A0A310SQ89 A0A026WVC2 A0A067QXX1 B3N5Q2 A0A195CG93 A0A0J9R3K2 B4HXN3 B4LV42 Q9VJP9 T1H9Y4 A0A2J7QZ13 A0A1W4VKP1 E2AG35 B4NYD0 A0A1B6J4Y7 K7JKY3 A0A023F9T7 A0A1B6LDL4 A0A0J7KFL9 E9ILP8 A0A0C9QWP7 A0A151XF60 A0A195DTJ5 B3MK60 E0W063 Q29KT7 F4WF82 A0A2M4AZ04 B4GSU2 A0A182ME64 A0A2P8YGC5 E2C992 A0A158NP47 T1DRW3 B4KLG1 A0A182RWJ8 A0A2M4AYU6 B4JQS7 A0A2S2Q8I3 A0A195F9J3 A0A3B0JHY8 A0A1B6CV90 A0A195AYS7 A0A2M4C2R3 A0A0M4ENR7 A0A182JBI8 A0A2M4C2Y4 A0A1B6F8T0 A0A2M4C3N5 A0A182T1W0 A0A182FA18 A0A2M4C2W6 A0A2M3ZDE7 B4MZ94 A0A2M4C2R5 U5ES69 A0A293LAA3 B0WAF8 A0A1B0G1N6 A0A1Q3FD04 A0A1A9VA21 A0A1A9ZLY9 A0A1S8VZ34 A0A0A9Y9X3 A0A336LXH0 A0A2R7X1K6 A0A182NG38 A0A084VTG7 H3A8W3 A0A3B1IER3 A0A1A9W7W2 W5JBC4 A0A1S8VYD1 A0A1A9XHL7 A0A1B0B1N6 Q7PSN0 A0A023EGT7 A0A1S3CWH2 A0A182HA06 W5UC07 A0A182TJK3 A0A3F2YSU2 A0A1L8DUU4 A0A1Y9IZX9
PDB
2ZDI
E-value=0.00869497,
Score=86
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
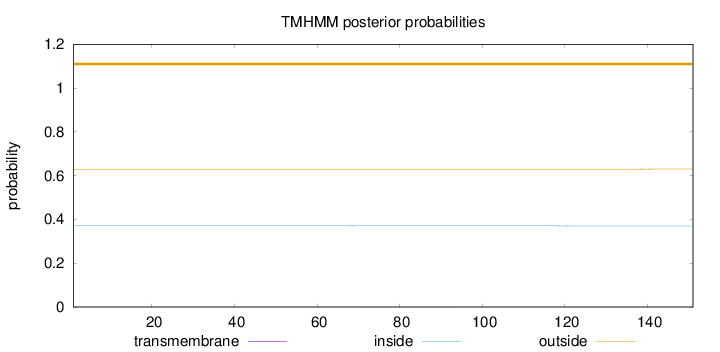
Length:
151
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01824
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.37082
outside
1 - 151
Population Genetic Test Statistics
Pi
189.031747
Theta
155.064169
Tajima's D
0.711522
CLR
0
CSRT
0.578671066446678
Interpretation
Uncertain
