Gene
KWMTBOMO15360
Pre Gene Modal
BGIBMGA005082
Annotation
PREDICTED:_potassium_channel_subfamily_K_member_9_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.85
Sequence
CDS
ATGAAGCGGCAGAATGTACGAACATTGTCACTTGTGGTTTGCACTTTTACGTATCTTCTCATCGGAGCAGCGGTCTTTGATGCATTGGAGTCAGACACCGAAAGTAAAAGATTAGAGGTGCTGTCAGACATGAAAAATGGCTTATTACGCAAATATAATATAAGTGCTGAAGACTACCATGTGATAGAAATAGTATTGATAGAGAATAAGCCCCACAAAGCTGGTCCACAGTGGAAGTTCGCAGGAGCATTCTATTTTGCCACTGTCGTTTTAGCTATGATTGGCTATGGACATTCCACACCTGTTACTGTAGGAGGAAAGGCTTTTTGTATGGCCTATGCCATGGTGGGTATACCCCTAGGGCTGGTGATGTTCCAGAGTATCGGGGAACGGTTAAATAAGTTCGCTTCAGTTGTGATTCGTAGAGCGAAATGTTATCTGCGTTGTAATACCACTGAAGCAACCGAAATGAATCTGATGTTTGCTACTGGAATGCTCTCATCAATCATAATAACTACAGGAGCCGCCGTATTCTCTCGGTACGAAGGCTGGAGCTACTTCGATAGTTTCTATTACTGCTTCGTGACGCTGACCACCATCGGCTTCGGAGACTACGTCGCTCTGCAAAACGATCAAGCCTTGACCAGTAAACCCGGATATGTGGCCTTGAGTCTCGTCTTCATATTGTTCGGGCTGGCCGTTGTCGCCGCCAGCATCAACTTACTCGTTCTCAGATTCATGACCATGCAAGCCGAAGAGAACGCTCGAGACGAAGACAAAGAAGGTTCTAGAACATTGCTACCGATAGAGGGACACGGCATCGCGGCCCGGCAGCGTTCACACGACGACCAGGCGTCTGTATGCTCGTGCAACTGTTTGGGCAATAAGCAGTGTGAGAGCGGAGCTTTACTCGAAGCTCCTACCCGACCTTATCGTCTGCGAGCGCGGGCGTCTCTCACTCAAGACATCATCGAACGAGCATCCGTTTAA
Protein
MKRQNVRTLSLVVCTFTYLLIGAAVFDALESDTESKRLEVLSDMKNGLLRKYNISAEDYHVIEIVLIENKPHKAGPQWKFAGAFYFATVVLAMIGYGHSTPVTVGGKAFCMAYAMVGIPLGLVMFQSIGERLNKFASVVIRRAKCYLRCNTTEATEMNLMFATGMLSSIIITTGAAVFSRYEGWSYFDSFYYCFVTLTTIGFGDYVALQNDQALTSKPGYVALSLVFILFGLAVVAASINLLVLRFMTMQAEENARDEDKEGSRTLLPIEGHGIAARQRSHDDQASVCSCNCLGNKQCESGALLEAPTRPYRLRARASLTQDIIERASV
Summary
Similarity
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Belongs to the tubulin family.
Belongs to the tubulin family.
Uniprot
A0A2A4J4L4
A0A2H1WFY8
A0A194RL23
A0A194Q7V6
A0A212FJ93
A0A2W1BDJ7
+ More
A0A2S1ZBF1 A0A1B6CEA1 K7IR70 A0A0M9A4P6 A0A0C9R6I7 A0A1I8MRJ4 A0A088A5T7 A0A067QGT3 A0A1A9YRD9 A0A1A9UYZ4 A0A1B0FFP9 A0A1B6L1K7 A0A1A9Z9N2 A0A154P5R5 A0A0K8UZP6 A0A1I8PKQ8 A0A2A3EKK6 A0A0A9XJ12 A0A195FB09 E2A4D4 A0A0A1XCQ6 A0A310SPY7 A0A151J9F5 A0A195CMP3 A0A151I2T9 A0A151XCK4 A0A1B0GIG7 A0A034VVK3 A0A026W797 A0A2S2Q7N3 A0A1S4FGL8 Q172H0 A0A182H1K4 A0A1A9WY63 A0A182IX12 A0A182XWW9 A0A084WRD3 A0A182PAE7 A0A182UZZ3 A0A182JRI2 A0A182UJZ1 Q7QC61 A0A182HSZ5 A0A182LU24 W5JL22 A0A182VXD8 A0A182FT44 A0A182RA10 B0WAG7 A0A1B0D5S1 D6WP56 A0A232EG34 Q9VHE0 Q3ZZY0 B3M058 B4PUN4 B4HKD9 B3NZS5 B4N8V9 B4GP60 A0A1W4VPF3 B4QWM6 B4JEL0 A0A3B0KC46 Q29BM1 A0A0L0CLP5 A0A2R7WII9 A0A0M3QYI7 B4LW19 A0A1J1HJW7 B4K4Z7 W8ANQ1 A0A0K8UYQ1 F4WYR4 A0A0P4VG60 E2BHQ9 A0A1W4XC12 A0A182SZK2 A0A182XI12 N6TPZ0 A0A1S3DH63 A0A0L7R457 A0A1D2NHZ0 A0A1B0CQ52 A0A0K8V6N9 A0A0K8URQ7 A0A1C9TAA8 A0A0P4W2I0 A0A1Q3FHU4 A0A2P8YPD0 A0A0P5JH38 A0A182QFP7 A0A0J7L252 E9G6Z1
A0A2S1ZBF1 A0A1B6CEA1 K7IR70 A0A0M9A4P6 A0A0C9R6I7 A0A1I8MRJ4 A0A088A5T7 A0A067QGT3 A0A1A9YRD9 A0A1A9UYZ4 A0A1B0FFP9 A0A1B6L1K7 A0A1A9Z9N2 A0A154P5R5 A0A0K8UZP6 A0A1I8PKQ8 A0A2A3EKK6 A0A0A9XJ12 A0A195FB09 E2A4D4 A0A0A1XCQ6 A0A310SPY7 A0A151J9F5 A0A195CMP3 A0A151I2T9 A0A151XCK4 A0A1B0GIG7 A0A034VVK3 A0A026W797 A0A2S2Q7N3 A0A1S4FGL8 Q172H0 A0A182H1K4 A0A1A9WY63 A0A182IX12 A0A182XWW9 A0A084WRD3 A0A182PAE7 A0A182UZZ3 A0A182JRI2 A0A182UJZ1 Q7QC61 A0A182HSZ5 A0A182LU24 W5JL22 A0A182VXD8 A0A182FT44 A0A182RA10 B0WAG7 A0A1B0D5S1 D6WP56 A0A232EG34 Q9VHE0 Q3ZZY0 B3M058 B4PUN4 B4HKD9 B3NZS5 B4N8V9 B4GP60 A0A1W4VPF3 B4QWM6 B4JEL0 A0A3B0KC46 Q29BM1 A0A0L0CLP5 A0A2R7WII9 A0A0M3QYI7 B4LW19 A0A1J1HJW7 B4K4Z7 W8ANQ1 A0A0K8UYQ1 F4WYR4 A0A0P4VG60 E2BHQ9 A0A1W4XC12 A0A182SZK2 A0A182XI12 N6TPZ0 A0A1S3DH63 A0A0L7R457 A0A1D2NHZ0 A0A1B0CQ52 A0A0K8V6N9 A0A0K8URQ7 A0A1C9TAA8 A0A0P4W2I0 A0A1Q3FHU4 A0A2P8YPD0 A0A0P5JH38 A0A182QFP7 A0A0J7L252 E9G6Z1
Pubmed
26354079
22118469
28756777
20075255
25315136
24845553
+ More
25401762 26823975 20798317 25830018 25348373 24508170 30249741 17510324 26483478 25244985 24438588 12364791 14747013 17210077 20920257 23761445 18362917 19820115 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17074048 17994087 17550304 15632085 26108605 24495485 21719571 27129103 23537049 27289101 29403074 21292972
25401762 26823975 20798317 25830018 25348373 24508170 30249741 17510324 26483478 25244985 24438588 12364791 14747013 17210077 20920257 23761445 18362917 19820115 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17074048 17994087 17550304 15632085 26108605 24495485 21719571 27129103 23537049 27289101 29403074 21292972
EMBL
NWSH01003255
PCG66676.1
ODYU01008408
SOQ51983.1
KQ460045
KPJ18025.1
+ More
KQ459324 KPJ01474.1 AGBW02008295 OWR53801.1 KZ150247 PZC71864.1 MF373406 AWK21960.1 GEDC01025583 JAS11715.1 KQ435756 KOX75883.1 GBYB01008421 GBYB01008422 GBYB01008423 JAG78188.1 JAG78189.1 JAG78190.1 KK853897 KDQ71519.1 CCAG010009890 GEBQ01022456 JAT17521.1 KQ434809 KZC06538.1 GDHF01022775 GDHF01020534 JAI29539.1 JAI31780.1 KZ288229 PBC31709.1 GBHO01023620 GDHC01000814 JAG19984.1 JAQ17815.1 KQ981727 KYN37219.1 GL436635 EFN71681.1 GBXI01005567 JAD08725.1 KQ760329 OAD60870.1 KQ979425 KYN21612.1 KQ977565 KYN01978.1 KQ976508 KYM82743.1 KQ982298 KYQ58126.1 AJWK01014097 GAKP01013082 JAC45870.1 KK107366 QOIP01000014 EZA51863.1 RLU15134.1 GGMS01004534 MBY73737.1 CH477437 EAT40927.1 JXUM01098717 JXUM01103756 KQ564831 KQ564442 KXJ71681.1 KXJ72260.1 ATLV01026020 KE525405 KFB52777.1 AAAB01008859 EAA08170.2 APCN01000376 AXCM01000493 ADMH02001133 ETN63998.1 DS231872 EDS41390.1 AJVK01025559 KQ971352 EFA07264.1 NNAY01004869 OXU17320.1 AE014297 BT024313 AAF54374.1 ABC86375.1 AJ920331 CAI72673.1 CH902617 EDV42015.1 CM000160 EDW96651.1 CH480815 EDW42889.1 CH954181 EDV49785.1 CH964232 EDW81560.2 CH479186 EDW38943.1 CM000364 EDX13630.1 CH916369 EDV93141.1 OUUW01000005 SPP81178.1 CM000070 EAL26976.3 JRES01000228 KNC33181.1 KK854883 PTY19467.1 CP012526 ALC47689.1 CH940650 EDW66524.2 CVRI01000006 CRK88335.1 CH933806 EDW13968.2 GAMC01018928 JAB87627.1 GDHF01020522 JAI31792.1 GL888463 EGI60568.1 GDKW01003464 JAI53131.1 GL448324 EFN84742.1 APGK01025935 KB740605 KB632095 ENN80063.1 ERL88733.1 KQ414658 KOC65633.1 LJIJ01000040 ODN04596.1 AJWK01023079 GDHF01017763 JAI34551.1 GDHF01022942 JAI29372.1 KU681449 AOR07230.1 GDRN01094209 JAI59748.1 GFDL01007927 JAV27118.1 PYGN01000452 PSN46117.1 GDIQ01202586 JAK49139.1 AXCN02000116 LBMM01001307 KMQ96539.1 GL732534 EFX84367.1
KQ459324 KPJ01474.1 AGBW02008295 OWR53801.1 KZ150247 PZC71864.1 MF373406 AWK21960.1 GEDC01025583 JAS11715.1 KQ435756 KOX75883.1 GBYB01008421 GBYB01008422 GBYB01008423 JAG78188.1 JAG78189.1 JAG78190.1 KK853897 KDQ71519.1 CCAG010009890 GEBQ01022456 JAT17521.1 KQ434809 KZC06538.1 GDHF01022775 GDHF01020534 JAI29539.1 JAI31780.1 KZ288229 PBC31709.1 GBHO01023620 GDHC01000814 JAG19984.1 JAQ17815.1 KQ981727 KYN37219.1 GL436635 EFN71681.1 GBXI01005567 JAD08725.1 KQ760329 OAD60870.1 KQ979425 KYN21612.1 KQ977565 KYN01978.1 KQ976508 KYM82743.1 KQ982298 KYQ58126.1 AJWK01014097 GAKP01013082 JAC45870.1 KK107366 QOIP01000014 EZA51863.1 RLU15134.1 GGMS01004534 MBY73737.1 CH477437 EAT40927.1 JXUM01098717 JXUM01103756 KQ564831 KQ564442 KXJ71681.1 KXJ72260.1 ATLV01026020 KE525405 KFB52777.1 AAAB01008859 EAA08170.2 APCN01000376 AXCM01000493 ADMH02001133 ETN63998.1 DS231872 EDS41390.1 AJVK01025559 KQ971352 EFA07264.1 NNAY01004869 OXU17320.1 AE014297 BT024313 AAF54374.1 ABC86375.1 AJ920331 CAI72673.1 CH902617 EDV42015.1 CM000160 EDW96651.1 CH480815 EDW42889.1 CH954181 EDV49785.1 CH964232 EDW81560.2 CH479186 EDW38943.1 CM000364 EDX13630.1 CH916369 EDV93141.1 OUUW01000005 SPP81178.1 CM000070 EAL26976.3 JRES01000228 KNC33181.1 KK854883 PTY19467.1 CP012526 ALC47689.1 CH940650 EDW66524.2 CVRI01000006 CRK88335.1 CH933806 EDW13968.2 GAMC01018928 JAB87627.1 GDHF01020522 JAI31792.1 GL888463 EGI60568.1 GDKW01003464 JAI53131.1 GL448324 EFN84742.1 APGK01025935 KB740605 KB632095 ENN80063.1 ERL88733.1 KQ414658 KOC65633.1 LJIJ01000040 ODN04596.1 AJWK01023079 GDHF01017763 JAI34551.1 GDHF01022942 JAI29372.1 KU681449 AOR07230.1 GDRN01094209 JAI59748.1 GFDL01007927 JAV27118.1 PYGN01000452 PSN46117.1 GDIQ01202586 JAK49139.1 AXCN02000116 LBMM01001307 KMQ96539.1 GL732534 EFX84367.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000002358
UP000053105
+ More
UP000095301 UP000005203 UP000027135 UP000092443 UP000078200 UP000092444 UP000092445 UP000076502 UP000095300 UP000242457 UP000078541 UP000000311 UP000078492 UP000078542 UP000078540 UP000075809 UP000092461 UP000053097 UP000279307 UP000008820 UP000069940 UP000249989 UP000091820 UP000075880 UP000076408 UP000030765 UP000075885 UP000075903 UP000075881 UP000075902 UP000007062 UP000075840 UP000075883 UP000000673 UP000075920 UP000069272 UP000075900 UP000002320 UP000092462 UP000007266 UP000215335 UP000000803 UP000007801 UP000002282 UP000001292 UP000008711 UP000007798 UP000008744 UP000192221 UP000000304 UP000001070 UP000268350 UP000001819 UP000037069 UP000092553 UP000008792 UP000183832 UP000009192 UP000007755 UP000008237 UP000192223 UP000075901 UP000076407 UP000019118 UP000030742 UP000079169 UP000053825 UP000094527 UP000245037 UP000075886 UP000036403 UP000000305
UP000095301 UP000005203 UP000027135 UP000092443 UP000078200 UP000092444 UP000092445 UP000076502 UP000095300 UP000242457 UP000078541 UP000000311 UP000078492 UP000078542 UP000078540 UP000075809 UP000092461 UP000053097 UP000279307 UP000008820 UP000069940 UP000249989 UP000091820 UP000075880 UP000076408 UP000030765 UP000075885 UP000075903 UP000075881 UP000075902 UP000007062 UP000075840 UP000075883 UP000000673 UP000075920 UP000069272 UP000075900 UP000002320 UP000092462 UP000007266 UP000215335 UP000000803 UP000007801 UP000002282 UP000001292 UP000008711 UP000007798 UP000008744 UP000192221 UP000000304 UP000001070 UP000268350 UP000001819 UP000037069 UP000092553 UP000008792 UP000183832 UP000009192 UP000007755 UP000008237 UP000192223 UP000075901 UP000076407 UP000019118 UP000030742 UP000079169 UP000053825 UP000094527 UP000245037 UP000075886 UP000036403 UP000000305
Interpro
IPR003280
2pore_dom_K_chnl
+ More
IPR013099 K_chnl_dom
IPR003092 2pore_dom_K_chnl_TASK
IPR002453 Beta_tubulin
IPR037103 Tubulin/FtsZ_C_sf
IPR008280 Tub_FtsZ_C
IPR003008 Tubulin_FtsZ_GTPase
IPR017975 Tubulin_CS
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR000217 Tubulin
IPR023123 Tubulin_C
IPR006876 LMBR1-like_membr_prot
IPR013099 K_chnl_dom
IPR003092 2pore_dom_K_chnl_TASK
IPR002453 Beta_tubulin
IPR037103 Tubulin/FtsZ_C_sf
IPR008280 Tub_FtsZ_C
IPR003008 Tubulin_FtsZ_GTPase
IPR017975 Tubulin_CS
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR000217 Tubulin
IPR023123 Tubulin_C
IPR006876 LMBR1-like_membr_prot
Gene 3D
ProteinModelPortal
A0A2A4J4L4
A0A2H1WFY8
A0A194RL23
A0A194Q7V6
A0A212FJ93
A0A2W1BDJ7
+ More
A0A2S1ZBF1 A0A1B6CEA1 K7IR70 A0A0M9A4P6 A0A0C9R6I7 A0A1I8MRJ4 A0A088A5T7 A0A067QGT3 A0A1A9YRD9 A0A1A9UYZ4 A0A1B0FFP9 A0A1B6L1K7 A0A1A9Z9N2 A0A154P5R5 A0A0K8UZP6 A0A1I8PKQ8 A0A2A3EKK6 A0A0A9XJ12 A0A195FB09 E2A4D4 A0A0A1XCQ6 A0A310SPY7 A0A151J9F5 A0A195CMP3 A0A151I2T9 A0A151XCK4 A0A1B0GIG7 A0A034VVK3 A0A026W797 A0A2S2Q7N3 A0A1S4FGL8 Q172H0 A0A182H1K4 A0A1A9WY63 A0A182IX12 A0A182XWW9 A0A084WRD3 A0A182PAE7 A0A182UZZ3 A0A182JRI2 A0A182UJZ1 Q7QC61 A0A182HSZ5 A0A182LU24 W5JL22 A0A182VXD8 A0A182FT44 A0A182RA10 B0WAG7 A0A1B0D5S1 D6WP56 A0A232EG34 Q9VHE0 Q3ZZY0 B3M058 B4PUN4 B4HKD9 B3NZS5 B4N8V9 B4GP60 A0A1W4VPF3 B4QWM6 B4JEL0 A0A3B0KC46 Q29BM1 A0A0L0CLP5 A0A2R7WII9 A0A0M3QYI7 B4LW19 A0A1J1HJW7 B4K4Z7 W8ANQ1 A0A0K8UYQ1 F4WYR4 A0A0P4VG60 E2BHQ9 A0A1W4XC12 A0A182SZK2 A0A182XI12 N6TPZ0 A0A1S3DH63 A0A0L7R457 A0A1D2NHZ0 A0A1B0CQ52 A0A0K8V6N9 A0A0K8URQ7 A0A1C9TAA8 A0A0P4W2I0 A0A1Q3FHU4 A0A2P8YPD0 A0A0P5JH38 A0A182QFP7 A0A0J7L252 E9G6Z1
A0A2S1ZBF1 A0A1B6CEA1 K7IR70 A0A0M9A4P6 A0A0C9R6I7 A0A1I8MRJ4 A0A088A5T7 A0A067QGT3 A0A1A9YRD9 A0A1A9UYZ4 A0A1B0FFP9 A0A1B6L1K7 A0A1A9Z9N2 A0A154P5R5 A0A0K8UZP6 A0A1I8PKQ8 A0A2A3EKK6 A0A0A9XJ12 A0A195FB09 E2A4D4 A0A0A1XCQ6 A0A310SPY7 A0A151J9F5 A0A195CMP3 A0A151I2T9 A0A151XCK4 A0A1B0GIG7 A0A034VVK3 A0A026W797 A0A2S2Q7N3 A0A1S4FGL8 Q172H0 A0A182H1K4 A0A1A9WY63 A0A182IX12 A0A182XWW9 A0A084WRD3 A0A182PAE7 A0A182UZZ3 A0A182JRI2 A0A182UJZ1 Q7QC61 A0A182HSZ5 A0A182LU24 W5JL22 A0A182VXD8 A0A182FT44 A0A182RA10 B0WAG7 A0A1B0D5S1 D6WP56 A0A232EG34 Q9VHE0 Q3ZZY0 B3M058 B4PUN4 B4HKD9 B3NZS5 B4N8V9 B4GP60 A0A1W4VPF3 B4QWM6 B4JEL0 A0A3B0KC46 Q29BM1 A0A0L0CLP5 A0A2R7WII9 A0A0M3QYI7 B4LW19 A0A1J1HJW7 B4K4Z7 W8ANQ1 A0A0K8UYQ1 F4WYR4 A0A0P4VG60 E2BHQ9 A0A1W4XC12 A0A182SZK2 A0A182XI12 N6TPZ0 A0A1S3DH63 A0A0L7R457 A0A1D2NHZ0 A0A1B0CQ52 A0A0K8V6N9 A0A0K8URQ7 A0A1C9TAA8 A0A0P4W2I0 A0A1Q3FHU4 A0A2P8YPD0 A0A0P5JH38 A0A182QFP7 A0A0J7L252 E9G6Z1
PDB
6CQ9
E-value=5.6308e-26,
Score=291
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
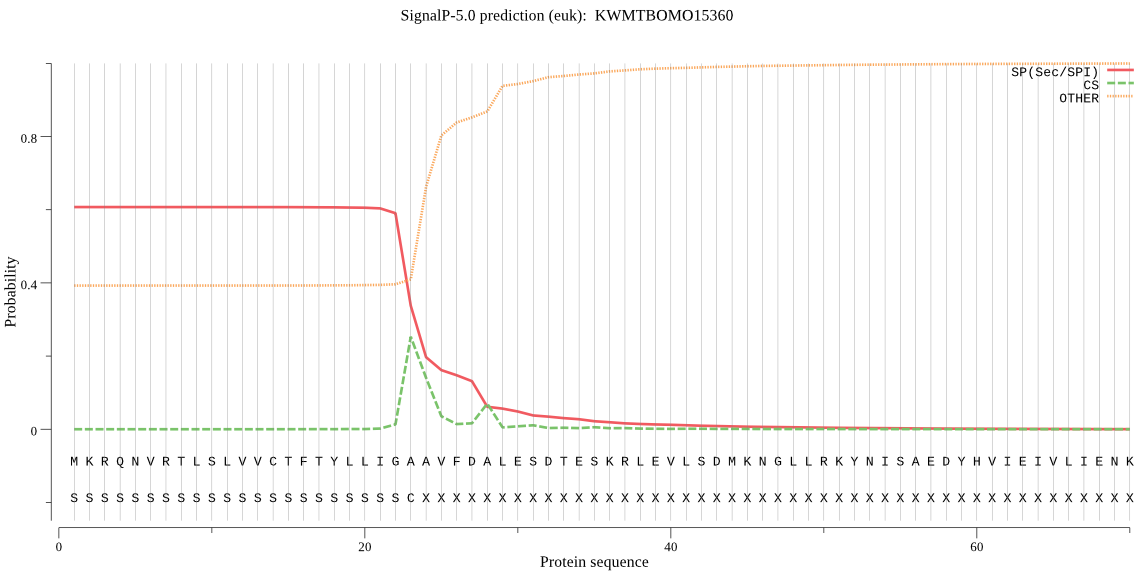
SignalP
Position: 1 - 23,
Likelihood: 0.607344
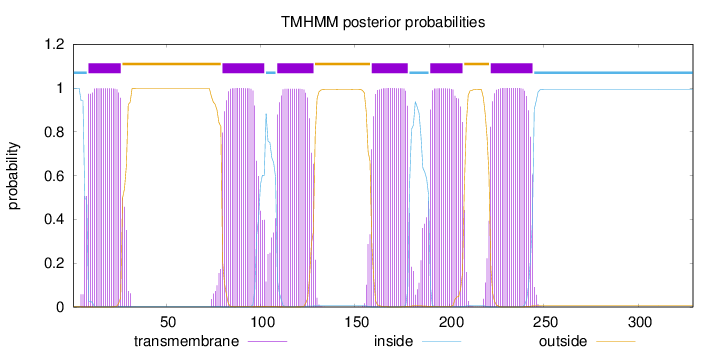
Length:
329
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
125.8867
Exp number, first 60 AAs:
20.40217
Total prob of N-in:
0.99905
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 26
outside
27 - 79
TMhelix
80 - 102
inside
103 - 108
TMhelix
109 - 128
outside
129 - 158
TMhelix
159 - 178
inside
179 - 189
TMhelix
190 - 207
outside
208 - 221
TMhelix
222 - 244
inside
245 - 329
Population Genetic Test Statistics
Pi
230.051346
Theta
156.33544
Tajima's D
0.873382
CLR
0.459751
CSRT
0.626218689065547
Interpretation
Uncertain
