Gene
KWMTBOMO15345
Pre Gene Modal
BGIBMGA005125
Annotation
DNApol-iota_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 4.019
Sequence
CDS
ATGGAATCGAGCTGTTCCACTTGTAACGAGATTGATGGTGAACATTTCAAGAGCATTATACATATTGATTTAGATTGTTTCTACGCGCAAGTGGAAATGGTGCGAAACCCCGAACTCCGTGATGTACCGCTCGGTATCCAACAGAAGAACATTGTAGTGACGAGTAATTACGAGGCAAGGAACTACGGAGTTAGTAAATGTATGCTCGTTACTGACGCACTTAAGGTTTGTCCAAATTTGAAACTTGTAAATGGTGAAGATTTACATAGTTACAGAACTGCATCAGGCAAGGTGTTTTTAGTGTTGCAAGGTTGGCGGTGTCCCGTTGAAAAACTTGGAATGGATGAAAATTTCATTGATGTGACAAGTCTTGTGGAGGAAAAATTAAAAAATGCAAACTTAAATGATCTCATAGTTATGGGCCAGCTATATAGTGAACCGAGTGCTGAATGTCCATGTGGTTGTCACAACAGACTAAAAATAGCATCACAAATTGCAAATGAAATGAGGAAGAAAATTTATGATGAGTTGGGTTACACAACTTGTGCCGGAATAGCTCATAACAAGTTGTTGGCTAAACTAATTTGTCCCCTGCATAAACCTAACAACCAGACCACAATATTTCCTCAACATGGAGCCAATTACATGTCTACTCTGACCAGTGTCCGGTCCATTCCAAGTATTGGCTCCAAAACAATGGAAGCATTAATTTCTCAGAAGATAATATCGGTCAGTGACTTACAAGAAGCTCCTTTAGAGATTCTGAAAAAACATTTCAGCAGTGATATGGCTGTTAGATTGAAAAGTTTAAGTTTAGGAGAGGATAATACTCCAGTAAAGCAAAGTGGAAAACCTCAGAGCATAGGATTGGAGGATAGTTTCAAAACTGTAAGTGTAAAAAGTGAGGTGGAGGATAAATTCTCTGCTCTGCTACAGAGATTATTAGTACTTGTAAGAGAAGATGGTCGCATTCCAGTTTCGGTGAGGGTAACTTTACGTAAGAAGGATGCTAAAAGATTGAGCAGTCATAGGGAATCAAGACAAAGTCAGATTTCTCCATCTATCTTCACACTAAGCAGTGGTGTGTTGACTGTAACAGAAGCTGGTCAACAAAAATTGATGGCCATAATAATGAGATTATTCACTAAACTTGTTGATTTATCTAAACCATTCCATTTGACTTTGGTGGGACTAGCGTTTACCAAGTTCCAAGAACGAATGACAGGCAGGAGTTCCATTGTAAACTACTTAATGAATGACATTTCTGTTCAGTCAGTTCTCAACTTACAAAATGATGGTGACGCTTCAATTACTTCAATGGACTATTCTGCTGCATCTCCAAGTAGCAGTACCACTACAGATCTCTCTGATGCAGAAATTGAGCCATCACCTAAAAAACCTAAGAAAGTTAATTGGATTGCAAAGAAGCGGTGCCTAGCCAAGGAAGAAGTTGCATCACCAAGTAAACTGAAAGTTGGTGAACTTAGACTTAATTCTAAAGAGCTAGAAAAAGTCTCAGAGTTGAGGTTAAATTCAAGAGATCGATCTCTTACTCCTCGAGCAAGCCCCGCTAAAGACAACTTTTCTGATGCTTCAGATTCTATGAAGGACCTAGAAGATGGAGCTTGTAATGATTGTCCAAGTTATGTTGATAAAGAAGTGTTTAGTGCTCTCCCTGATGACATGCAACAGGAACTTAAATCAATGTGGAAGAACCCTACTAGTTCTGAGAAAACCAGAAGTAGTCCAAGAAAGAACAAATCAAAACCCAACTCTATAATGAAATATTTTGTTCCACAAAAATAG
Protein
MESSCSTCNEIDGEHFKSIIHIDLDCFYAQVEMVRNPELRDVPLGIQQKNIVVTSNYEARNYGVSKCMLVTDALKVCPNLKLVNGEDLHSYRTASGKVFLVLQGWRCPVEKLGMDENFIDVTSLVEEKLKNANLNDLIVMGQLYSEPSAECPCGCHNRLKIASQIANEMRKKIYDELGYTTCAGIAHNKLLAKLICPLHKPNNQTTIFPQHGANYMSTLTSVRSIPSIGSKTMEALISQKIISVSDLQEAPLEILKKHFSSDMAVRLKSLSLGEDNTPVKQSGKPQSIGLEDSFKTVSVKSEVEDKFSALLQRLLVLVREDGRIPVSVRVTLRKKDAKRLSSHRESRQSQISPSIFTLSSGVLTVTEAGQQKLMAIIMRLFTKLVDLSKPFHLTLVGLAFTKFQERMTGRSSIVNYLMNDISVQSVLNLQNDGDASITSMDYSAASPSSSTTTDLSDAEIEPSPKKPKKVNWIAKKRCLAKEEVASPSKLKVGELRLNSKELEKVSELRLNSRDRSLTPRASPAKDNFSDASDSMKDLEDGACNDCPSYVDKEVFSALPDDMQQELKSMWKNPTSSEKTRSSPRKNKSKPNSIMKYFVPQK
Summary
Uniprot
H9J6I3
A0A0L7KZA2
A0A2W1B7C9
A0A2A4ISL6
S4PY61
A0A2H1WQY3
+ More
A0A212EZQ1 A0A0N0PBG9 A0A194Q7X2 A0A1Y1MM18 A0A2Z5U5Y4 A0A067QRT1 A0A1B0CMT6 A0A1L8DWK2 A0A1L8DX51 A0A1B0GMD8 A0A0T6B000 A0A2J7QAX6 A0A154P3W2 A0A0N0BH96 B0XFI8 A0A2J7QAX1 A0A1Q3EXE3 A0A2A3EJ27 A0A026WSW5 E9J8Q1 A0A0L7R458 A0A310SHX1 A0A087ZQ84 Q175B7 A0A1S4FEH5 D6WQ70 A0A0J7KCT7 A0A195F1D5 A0A182GUP6 A0A182J948 A0A151XG28 F4WAV2 A0A151IH75 A0A195BWZ1 A0A158P3L5 A0A232FD27 B4NJ96 A0A1J1HUM8 A0A151ITB9 A0A2M4CIM7 A0A2M4CIB5 A0A2M4CIG5 A0A182FST1 B3LZW8 A0A1B6LND2 E2BNV5 A0A182SUQ7 K7J498 A0A182W2R3 A0A182RYA4 B4HLP4 A0A182XVI2 A0A182LV96 A0A034W883 A0A1A9VDQ2 B3NZW5 Q9VHV1 B4PSM0 A0A1W4W3C8 A0A1B0B2Z9 A0A240SWM0 A0A1A9ZSU6 A0A1B0FL96 B4K963 B4JF50 A0A0Q9X6P9 A0A182QW71 A0A0A1XEP7 A0A0M4ED21 A0A0L0BY70 A0A1A9W5P7 A0A1Q3EXL0 Q299A7 B4G4Y8 T1PJF1 B4M161 A0A1I8M628 A0A3B0JL79 A0A182JQC9 A0A1I8NLT7 A0A1I8NLU4 A0A182P580 A0A182XE77 A0A182TYG8 A0A182HJ46 A0A182LCA7 Q7QBI8 W8C8V8
A0A212EZQ1 A0A0N0PBG9 A0A194Q7X2 A0A1Y1MM18 A0A2Z5U5Y4 A0A067QRT1 A0A1B0CMT6 A0A1L8DWK2 A0A1L8DX51 A0A1B0GMD8 A0A0T6B000 A0A2J7QAX6 A0A154P3W2 A0A0N0BH96 B0XFI8 A0A2J7QAX1 A0A1Q3EXE3 A0A2A3EJ27 A0A026WSW5 E9J8Q1 A0A0L7R458 A0A310SHX1 A0A087ZQ84 Q175B7 A0A1S4FEH5 D6WQ70 A0A0J7KCT7 A0A195F1D5 A0A182GUP6 A0A182J948 A0A151XG28 F4WAV2 A0A151IH75 A0A195BWZ1 A0A158P3L5 A0A232FD27 B4NJ96 A0A1J1HUM8 A0A151ITB9 A0A2M4CIM7 A0A2M4CIB5 A0A2M4CIG5 A0A182FST1 B3LZW8 A0A1B6LND2 E2BNV5 A0A182SUQ7 K7J498 A0A182W2R3 A0A182RYA4 B4HLP4 A0A182XVI2 A0A182LV96 A0A034W883 A0A1A9VDQ2 B3NZW5 Q9VHV1 B4PSM0 A0A1W4W3C8 A0A1B0B2Z9 A0A240SWM0 A0A1A9ZSU6 A0A1B0FL96 B4K963 B4JF50 A0A0Q9X6P9 A0A182QW71 A0A0A1XEP7 A0A0M4ED21 A0A0L0BY70 A0A1A9W5P7 A0A1Q3EXL0 Q299A7 B4G4Y8 T1PJF1 B4M161 A0A1I8M628 A0A3B0JL79 A0A182JQC9 A0A1I8NLT7 A0A1I8NLU4 A0A182P580 A0A182XE77 A0A182TYG8 A0A182HJ46 A0A182LCA7 Q7QBI8 W8C8V8
Pubmed
19121390
26227816
28756777
23622113
22118469
26354079
+ More
28004739 26760975 24845553 24508170 30249741 21282665 17510324 18362917 19820115 26483478 21719571 21347285 28648823 17994087 20798317 20075255 25244985 25348373 10731132 11297519 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 25830018 26108605 15632085 23185243 25315136 20966253 12364791 14747013 17210077 24495485
28004739 26760975 24845553 24508170 30249741 21282665 17510324 18362917 19820115 26483478 21719571 21347285 28648823 17994087 20798317 20075255 25244985 25348373 10731132 11297519 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 25830018 26108605 15632085 23185243 25315136 20966253 12364791 14747013 17210077 24495485
EMBL
BABH01019770
JTDY01004191
KOB68475.1
KZ150250
PZC71829.1
NWSH01007896
+ More
PCG62765.1 GAIX01003848 JAA88712.1 ODYU01010346 SOQ55386.1 AGBW02011239 OWR46986.1 KQ460973 KPJ10056.1 KQ459324 KPJ01489.1 GEZM01030476 GEZM01030474 JAV85525.1 FX985786 BBA93673.1 KK853013 KDR12521.1 AJWK01019157 GFDF01003258 JAV10826.1 GFDF01003259 JAV10825.1 AJVK01024438 AJVK01024439 LJIG01016417 KRT80664.1 NEVH01016310 PNF25732.1 KQ434809 KZC06606.1 KQ435756 KOX75918.1 DS232930 EDS26826.1 PNF25733.1 GFDL01015085 JAV19960.1 KZ288229 PBC31727.1 KK107109 QOIP01000008 EZA59135.1 RLU19793.1 GL769036 EFZ10839.1 KQ414658 KOC65662.1 KQ759887 OAD62130.1 CH477403 EAT41670.1 KQ971354 EFA06118.2 LBMM01009440 KMQ88137.1 KQ981880 KYN33914.1 JXUM01089599 JXUM01089600 KQ982174 KYQ59317.1 GL888053 EGI68708.1 KQ977636 KYN01158.1 KQ976394 KYM93149.1 ADTU01008200 NNAY01000441 OXU28369.1 CH964272 EDW84927.1 CVRI01000021 CRK91775.1 KQ981039 KYN10141.1 GGFL01000917 MBW65095.1 GGFL01000914 MBW65092.1 GGFL01000915 MBW65093.1 CH902617 EDV44158.1 GEBQ01014898 JAT25079.1 GL449507 EFN82638.1 AAZX01006624 AAZX01007448 CH480815 EDW43071.1 AXCM01000024 GAKP01008034 GAKP01008033 JAC50919.1 CH954181 EDV49963.1 AE014297 AY051762 AB049434 AAF54198.1 AAK93186.1 AAN14307.1 AHN57226.1 BAB15800.1 CM000160 EDW96480.1 KRK03171.1 JXJN01007738 CCAG010013281 CH933806 EDW14476.1 KRG01029.1 CH916369 EDV93331.1 KRG01028.1 AXCN02001613 GBXI01015438 GBXI01004855 JAC98853.1 JAD09437.1 CP012526 ALC45628.1 JRES01001160 KNC24977.1 GFDL01015107 JAV19938.1 CM000070 EAL27796.1 KRT00292.1 CH479179 EDW24654.1 KA648240 AFP62869.1 CH940650 EDW67472.1 OUUW01000007 SPP83074.1 APCN01002124 AAAB01008879 EAA08441.5 GAMC01002709 GAMC01002708 JAC03847.1
PCG62765.1 GAIX01003848 JAA88712.1 ODYU01010346 SOQ55386.1 AGBW02011239 OWR46986.1 KQ460973 KPJ10056.1 KQ459324 KPJ01489.1 GEZM01030476 GEZM01030474 JAV85525.1 FX985786 BBA93673.1 KK853013 KDR12521.1 AJWK01019157 GFDF01003258 JAV10826.1 GFDF01003259 JAV10825.1 AJVK01024438 AJVK01024439 LJIG01016417 KRT80664.1 NEVH01016310 PNF25732.1 KQ434809 KZC06606.1 KQ435756 KOX75918.1 DS232930 EDS26826.1 PNF25733.1 GFDL01015085 JAV19960.1 KZ288229 PBC31727.1 KK107109 QOIP01000008 EZA59135.1 RLU19793.1 GL769036 EFZ10839.1 KQ414658 KOC65662.1 KQ759887 OAD62130.1 CH477403 EAT41670.1 KQ971354 EFA06118.2 LBMM01009440 KMQ88137.1 KQ981880 KYN33914.1 JXUM01089599 JXUM01089600 KQ982174 KYQ59317.1 GL888053 EGI68708.1 KQ977636 KYN01158.1 KQ976394 KYM93149.1 ADTU01008200 NNAY01000441 OXU28369.1 CH964272 EDW84927.1 CVRI01000021 CRK91775.1 KQ981039 KYN10141.1 GGFL01000917 MBW65095.1 GGFL01000914 MBW65092.1 GGFL01000915 MBW65093.1 CH902617 EDV44158.1 GEBQ01014898 JAT25079.1 GL449507 EFN82638.1 AAZX01006624 AAZX01007448 CH480815 EDW43071.1 AXCM01000024 GAKP01008034 GAKP01008033 JAC50919.1 CH954181 EDV49963.1 AE014297 AY051762 AB049434 AAF54198.1 AAK93186.1 AAN14307.1 AHN57226.1 BAB15800.1 CM000160 EDW96480.1 KRK03171.1 JXJN01007738 CCAG010013281 CH933806 EDW14476.1 KRG01029.1 CH916369 EDV93331.1 KRG01028.1 AXCN02001613 GBXI01015438 GBXI01004855 JAC98853.1 JAD09437.1 CP012526 ALC45628.1 JRES01001160 KNC24977.1 GFDL01015107 JAV19938.1 CM000070 EAL27796.1 KRT00292.1 CH479179 EDW24654.1 KA648240 AFP62869.1 CH940650 EDW67472.1 OUUW01000007 SPP83074.1 APCN01002124 AAAB01008879 EAA08441.5 GAMC01002709 GAMC01002708 JAC03847.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000027135 UP000092461 UP000092462 UP000235965 UP000076502 UP000053105 UP000002320 UP000242457 UP000053097 UP000279307 UP000053825 UP000005203 UP000008820 UP000007266 UP000036403 UP000078541 UP000069940 UP000075880 UP000075809 UP000007755 UP000078542 UP000078540 UP000005205 UP000215335 UP000007798 UP000183832 UP000078492 UP000069272 UP000007801 UP000008237 UP000075901 UP000002358 UP000075920 UP000075900 UP000001292 UP000076408 UP000075883 UP000078200 UP000008711 UP000000803 UP000002282 UP000192221 UP000092460 UP000092443 UP000092445 UP000092444 UP000009192 UP000001070 UP000075886 UP000092553 UP000037069 UP000091820 UP000001819 UP000008744 UP000008792 UP000095301 UP000268350 UP000075881 UP000095300 UP000075885 UP000076407 UP000075902 UP000075840 UP000075882 UP000007062
UP000027135 UP000092461 UP000092462 UP000235965 UP000076502 UP000053105 UP000002320 UP000242457 UP000053097 UP000279307 UP000053825 UP000005203 UP000008820 UP000007266 UP000036403 UP000078541 UP000069940 UP000075880 UP000075809 UP000007755 UP000078542 UP000078540 UP000005205 UP000215335 UP000007798 UP000183832 UP000078492 UP000069272 UP000007801 UP000008237 UP000075901 UP000002358 UP000075920 UP000075900 UP000001292 UP000076408 UP000075883 UP000078200 UP000008711 UP000000803 UP000002282 UP000192221 UP000092460 UP000092443 UP000092445 UP000092444 UP000009192 UP000001070 UP000075886 UP000092553 UP000037069 UP000091820 UP000001819 UP000008744 UP000008792 UP000095301 UP000268350 UP000075881 UP000095300 UP000075885 UP000076407 UP000075902 UP000075840 UP000075882 UP000007062
Interpro
SUPFAM
SSF100879
SSF100879
Gene 3D
ProteinModelPortal
H9J6I3
A0A0L7KZA2
A0A2W1B7C9
A0A2A4ISL6
S4PY61
A0A2H1WQY3
+ More
A0A212EZQ1 A0A0N0PBG9 A0A194Q7X2 A0A1Y1MM18 A0A2Z5U5Y4 A0A067QRT1 A0A1B0CMT6 A0A1L8DWK2 A0A1L8DX51 A0A1B0GMD8 A0A0T6B000 A0A2J7QAX6 A0A154P3W2 A0A0N0BH96 B0XFI8 A0A2J7QAX1 A0A1Q3EXE3 A0A2A3EJ27 A0A026WSW5 E9J8Q1 A0A0L7R458 A0A310SHX1 A0A087ZQ84 Q175B7 A0A1S4FEH5 D6WQ70 A0A0J7KCT7 A0A195F1D5 A0A182GUP6 A0A182J948 A0A151XG28 F4WAV2 A0A151IH75 A0A195BWZ1 A0A158P3L5 A0A232FD27 B4NJ96 A0A1J1HUM8 A0A151ITB9 A0A2M4CIM7 A0A2M4CIB5 A0A2M4CIG5 A0A182FST1 B3LZW8 A0A1B6LND2 E2BNV5 A0A182SUQ7 K7J498 A0A182W2R3 A0A182RYA4 B4HLP4 A0A182XVI2 A0A182LV96 A0A034W883 A0A1A9VDQ2 B3NZW5 Q9VHV1 B4PSM0 A0A1W4W3C8 A0A1B0B2Z9 A0A240SWM0 A0A1A9ZSU6 A0A1B0FL96 B4K963 B4JF50 A0A0Q9X6P9 A0A182QW71 A0A0A1XEP7 A0A0M4ED21 A0A0L0BY70 A0A1A9W5P7 A0A1Q3EXL0 Q299A7 B4G4Y8 T1PJF1 B4M161 A0A1I8M628 A0A3B0JL79 A0A182JQC9 A0A1I8NLT7 A0A1I8NLU4 A0A182P580 A0A182XE77 A0A182TYG8 A0A182HJ46 A0A182LCA7 Q7QBI8 W8C8V8
A0A212EZQ1 A0A0N0PBG9 A0A194Q7X2 A0A1Y1MM18 A0A2Z5U5Y4 A0A067QRT1 A0A1B0CMT6 A0A1L8DWK2 A0A1L8DX51 A0A1B0GMD8 A0A0T6B000 A0A2J7QAX6 A0A154P3W2 A0A0N0BH96 B0XFI8 A0A2J7QAX1 A0A1Q3EXE3 A0A2A3EJ27 A0A026WSW5 E9J8Q1 A0A0L7R458 A0A310SHX1 A0A087ZQ84 Q175B7 A0A1S4FEH5 D6WQ70 A0A0J7KCT7 A0A195F1D5 A0A182GUP6 A0A182J948 A0A151XG28 F4WAV2 A0A151IH75 A0A195BWZ1 A0A158P3L5 A0A232FD27 B4NJ96 A0A1J1HUM8 A0A151ITB9 A0A2M4CIM7 A0A2M4CIB5 A0A2M4CIG5 A0A182FST1 B3LZW8 A0A1B6LND2 E2BNV5 A0A182SUQ7 K7J498 A0A182W2R3 A0A182RYA4 B4HLP4 A0A182XVI2 A0A182LV96 A0A034W883 A0A1A9VDQ2 B3NZW5 Q9VHV1 B4PSM0 A0A1W4W3C8 A0A1B0B2Z9 A0A240SWM0 A0A1A9ZSU6 A0A1B0FL96 B4K963 B4JF50 A0A0Q9X6P9 A0A182QW71 A0A0A1XEP7 A0A0M4ED21 A0A0L0BY70 A0A1A9W5P7 A0A1Q3EXL0 Q299A7 B4G4Y8 T1PJF1 B4M161 A0A1I8M628 A0A3B0JL79 A0A182JQC9 A0A1I8NLT7 A0A1I8NLU4 A0A182P580 A0A182XE77 A0A182TYG8 A0A182HJ46 A0A182LCA7 Q7QBI8 W8C8V8
PDB
5KT7
E-value=1.44476e-83,
Score=790
Ontologies
GO
Topology
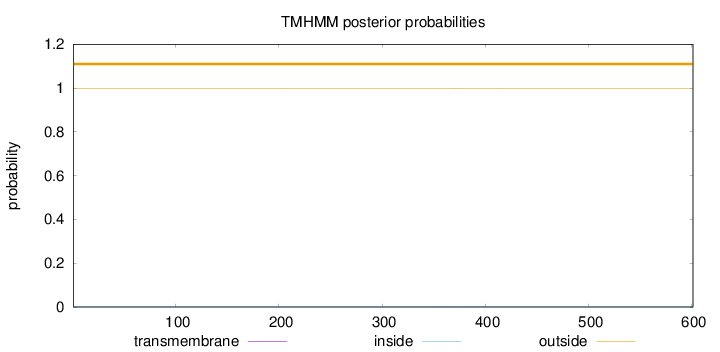
Length:
601
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01622
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00183
outside
1 - 601
Population Genetic Test Statistics
Pi
48.80834
Theta
18.705044
Tajima's D
-1.030711
CLR
708.352947
CSRT
0.137243137843108
Interpretation
Uncertain
