Gene
KWMTBOMO15342
Pre Gene Modal
BGIBMGA005075
Annotation
PREDICTED:_spastin_isoform_X3_[Bombyx_mori]
Full name
Spastin
Alternative Name
D-Spastin
Dm-Spastin
Dspastin
Dm-Spastin
Dspastin
Location in the cell
Mitochondrial Reliability : 1.444 Nuclear Reliability : 1.824
Sequence
CDS
ATGAAAAAGCCTCGTAAGATCGGCGAGTGCAACTTAGAAGTTGTAGTGAAAGACGGTATAGTGTCCACAGAGATAGCTCAAGAAGATATGTCTCATCACGTCGGACCAGGCGATCCACTGCTGGCCAAGCAGAAGCATCATCATCGCAAAGCTTTTGAATACATATCGAAAGCTTTGAAGATTGATGAGGAAAACGAAGGCCAAAAGGAGCTAGCAATAGAACTGTACAAGAAAGGGATTTTTGAATTGGAGCGAGGTATCGCTGTCGACTGCTGGGGAGGTAGAGGAGACGCTTGGCAAAGAGCTCAAAAACTACACGACAAGATGAAAACAAACCTTGGAATGGCTAAGGATCGTTTACATTTCCTTGTTGCAGGACGAAAACTGACGACGGCAGGCCGAAGAGTGCCTAGCAGTGGAGGAGGGCCCTTGATGAAATCTCAGACTTTGCCGAGGTCCATGGGCAGATCATCCTCCCAGCCGAACAGCTCTAACGGGGGCTACACACGGTACCCCATGAAGCCTGCGTCTACACCGCCGGCTGTTAAAAGACAGTTGTCTGTGCCGGTAAACGGTTCGCCAGTCCGCCGGGCGGTGAGTGGCTCGCAACGCGGTACGCCCACCAGAAGCAGGACTCCGCAGCCCGCGCTAACCGTCCGAGGCGTCGACCCGAAGCTGGTCCAGCTTATACTGGACGAGATAGTCGAAGGAGGACCCAAAGTAAACTGGGACGATATAGCTGGCCAAGAGGCAGCAAAACAAGCATTGCAAGAAATGGTTGTTCTACCTTCGTTGAGGCCCGAACTCTTCACCGGACTCCGATCGCCAGCCCGAGGCTTACTTTTGTTTGGACCTCCAGGAAATGGCAAGACTCTGCTAGCGCGATGCGTAGCGGCCGAATGCTCAGCGACTTTCTTCTCGATATCCGCCGCGAGTCTGACCAGCAAATATGTGGGCGAAGGCGAGAAGATGGTGAGGGCTCTGTTCCAAGTCGCTAGAGAACTGCAGCCTTCAATAATATTCGTGGATGAGGTAGACTCGCTGCTGTGCGAGAGATCGTCCGGCGAGCACGAGGCGTCTCGGCGGCTGAAGACAGAGTTCCTGGTGGAATTCGACGGACTCCCGGCGGCCGGAGCCGACAGACTCATCGTGATGGCCGCCACGAACCGACCTCAGGAACTGGACGAGGCTGCGTTGAGACGGTTCCCGAAGCGCGTGTACGTGTCGCTGCCGGACGCGCGGGTGCGGGCGGCGCTGGTGCGCGGCGTGCTGGCCCGCGGCGCCGCGCAGACCGAGCTCGCGGACGACGAGCTGGTGCGCCTCGCCGCGCTCACTGACGGCTACTCCGGCAGCGACCTCACCGCGCTCTGCAGGGACGCCGCGCTCGGCCCCATACGAGAGCTGGATCCCGAGGAAGTCAAATGTCTGGACCTGTCATTGGTCCGGAGCATTACCTTCCAAGATTTCCTAGATTCGCTGAAAAGGATCCGACCTTCTGTGTCTCCGCACAGCCTCGTCGCGTACGAGAAGTGGTCCGTGCAGTATGGCGATTTGGGATTTTGA
Protein
MKKPRKIGECNLEVVVKDGIVSTEIAQEDMSHHVGPGDPLLAKQKHHHRKAFEYISKALKIDEENEGQKELAIELYKKGIFELERGIAVDCWGGRGDAWQRAQKLHDKMKTNLGMAKDRLHFLVAGRKLTTAGRRVPSSGGGPLMKSQTLPRSMGRSSSQPNSSNGGYTRYPMKPASTPPAVKRQLSVPVNGSPVRRAVSGSQRGTPTRSRTPQPALTVRGVDPKLVQLILDEIVEGGPKVNWDDIAGQEAAKQALQEMVVLPSLRPELFTGLRSPARGLLLFGPPGNGKTLLARCVAAECSATFFSISAASLTSKYVGEGEKMVRALFQVARELQPSIIFVDEVDSLLCERSSGEHEASRRLKTEFLVEFDGLPAAGADRLIVMAATNRPQELDEAALRRFPKRVYVSLPDARVRAALVRGVLARGAAQTELADDELVRLAALTDGYSGSDLTALCRDAALGPIRELDPEEVKCLDLSLVRSITFQDFLDSLKRIRPSVSPHSLVAYEKWSVQYGDLGF
Summary
Description
ATP-dependent microtubule severing protein. Microtubule severing may promote reorganization of cellular microtubule arrays and the release of microtubules from the microtubule organizing center following nucleation.
ATP-dependent microtubule severing protein. Stimulates microtubule minus-end depolymerization and poleward microtubule flux in the mitotic spindle. Regulates microtubule stability in the neuromuscular junction synapse. Involved in lipid metabolism by regulating the size and distribution of lipid droplets. Involved in axon regeneration by regulating microtubule severing.
ATP-dependent microtubule severing protein. Stimulates microtubule minus-end depolymerization and poleward microtubule flux in the mitotic spindle (PubMed:15242610, PubMed:15562320, PubMed:15823537, PubMed:16276413, PubMed:17452528, PubMed:25875445, PubMed:18202664, PubMed:19341724). Regulates microtubule stability in the neuromuscular junction synapse (PubMed:15242610, PubMed:15562320, PubMed:19341724). Involved in lipid metabolism by regulating the size and distribution of lipid droplets (PubMed:25875445). Involved in axon regeneration by regulating microtubule severing (PubMed:23122959).
ATP-dependent microtubule severing protein that specifically recognizes and cuts microtubules that are polyglutamylated. Preferentially recognizes and acts on microtubules decorated with short polyglutamate tails: severing activity increases as the number of glutamates per tubulin rises from one to eight, but decreases beyond this glutamylation threshold. Microtubule severing promotes reorganization of cellular microtubule arrays and the release of microtubules from the centrosome following nucleation. Required for membrane traffic from the endoplasmic reticulum (ER) to the Golgi and for completion of the abscission stage of cytokinesis. Also plays a role in axon growth and the formation of axonal branches.
ATP-dependent microtubule severing protein. Stimulates microtubule minus-end depolymerization and poleward microtubule flux in the mitotic spindle. Regulates microtubule stability in the neuromuscular junction synapse. Involved in lipid metabolism by regulating the size and distribution of lipid droplets. Involved in axon regeneration by regulating microtubule severing.
ATP-dependent microtubule severing protein. Stimulates microtubule minus-end depolymerization and poleward microtubule flux in the mitotic spindle (PubMed:15242610, PubMed:15562320, PubMed:15823537, PubMed:16276413, PubMed:17452528, PubMed:25875445, PubMed:18202664, PubMed:19341724). Regulates microtubule stability in the neuromuscular junction synapse (PubMed:15242610, PubMed:15562320, PubMed:19341724). Involved in lipid metabolism by regulating the size and distribution of lipid droplets (PubMed:25875445). Involved in axon regeneration by regulating microtubule severing (PubMed:23122959).
ATP-dependent microtubule severing protein that specifically recognizes and cuts microtubules that are polyglutamylated. Preferentially recognizes and acts on microtubules decorated with short polyglutamate tails: severing activity increases as the number of glutamates per tubulin rises from one to eight, but decreases beyond this glutamylation threshold. Microtubule severing promotes reorganization of cellular microtubule arrays and the release of microtubules from the centrosome following nucleation. Required for membrane traffic from the endoplasmic reticulum (ER) to the Golgi and for completion of the abscission stage of cytokinesis. Also plays a role in axon growth and the formation of axonal branches.
Catalytic Activity
n ATP + n H(2)O + a microtubule = n ADP + n phosphate + (n+1) alpha/beta tubulin heterodimers.
Subunit
Homohexamer. The homohexamer is stabilized by ATP-binding. The homohexamer may adopt a ring conformation through which microtubules pass prior to being severed. Interacts with microtubules.
Homohexamer. The homohexamer is stabilized by ATP-binding. The homohexamer may adopt a ring conformation through which microtubules pass prior to being severed. Interacts with microtubules. Interacts with atl; may be involved in microtubule dynamics.
Homohexamer. The homohexamer is stabilized by ATP-binding. The homohexamer may adopt a ring conformation through which microtubules pass prior to being severed (PubMed:18202664). Interacts with microtubules (PubMed:15823537, PubMed:18202664). Interacts with atl; may be involved in microtubule dynamics (PubMed:19341724).
Homohexamer. The homohexamer is stabilized by ATP-binding. The homohexamer may adopt a ring conformation through which microtubules pass prior to being severed. Interacts with microtubules. Interacts with atl; may be involved in microtubule dynamics.
Homohexamer. The homohexamer is stabilized by ATP-binding. The homohexamer may adopt a ring conformation through which microtubules pass prior to being severed (PubMed:18202664). Interacts with microtubules (PubMed:15823537, PubMed:18202664). Interacts with atl; may be involved in microtubule dynamics (PubMed:19341724).
Similarity
Belongs to the AAA ATPase family. Spastin subfamily.
Belongs to the AAA ATPase family.
Belongs to the AAA ATPase family.
Keywords
ATP-binding
Cell cycle
Cell division
Chromosome
Complete proteome
Cytoplasm
Cytoskeleton
Developmental protein
Differentiation
Isomerase
Lipid droplet
Membrane
Microtubule
Mitosis
Neurogenesis
Nucleotide-binding
Reference proteome
3D-structure
Phosphoprotein
Alternative splicing
Nucleus
Feature
chain Spastin
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4IUU0
A0A194Q7H6
A0A2A4IU74
A0A0N1I5K3
H9J6D3
A0A212EZS7
+ More
A0A2W1B7L8 A0A1Y1KFA1 A0A1Y1KL42 A0A158NSN8 A0A1Y1KHQ5 A0A1Y1KMR0 A0A1Q3EUM0 A0A1Q3EUZ1 D6WR93 N6SRM3 A0A3B0JKI5 A0A034W3J6 A0A1W4U7W3 A0A0K8UYH1 A0A0R3NH26 A0A0A1WSU7 A0A1L8DU08 A0A1L8DTP0 A0A1I8NNF4 E0VYI4 A0A0M4F6S0 W8C8D8 A0A0Q9WIN0 A0A1W4W830 A0A0P8XY81 A0A0Q9WTF8 A0A1B6I365 A0A0Q9WY76 A0A1W4ULF9 A0A126GV13 A0A0B4LHJ5 A0A146L6H0 A0A126GV10 A0A0A9ZEL2 A0A0A9ZIL4 B4HGG6 Q8I0P1 A0A1B6KIZ5 A0A088A016 B4QSF0 F4W8A4 E2AI24 A0A195CAZ2 A0A1B6GJT6 A0A069DV18 A0A0V0G8F5 A0A224XLX6 A0A0P4VTL2 A0A182P867 A0A1B0D797 A0A0P4W909 A0A3Q0JEE6 A0A2J7Q240 A0A1B6DYR9 R7TU42 A0A2R5L4N1 A0A0L7R7C7 E2BI57 A0A2A3E4B6 A0A3Q2E8P7 A0A3L8DA27 A0A026WUX2 A0A151I5Z7 A0A195DUQ1 A0A154P359 A0A1B6CUX1 A0A1Z5KY73 A0A2L2YK85 A0A0C9PQF2 A0A151XDP6 A0A0T6B521 A0A2J7Q222 A0A2J7Q234 A0A0C9QYQ7 A0A3P8UBX3 A0A3Q1CK62 A0A1B6G657 A0A146MUH2 A0A3Q2TUE9 A0A3B3YLW5 A0A3Q1EVP2 M3ZW57 A0A1L1RP85 A0A146RUZ8 A0A2M4BN30 A0A1V4JL95 A0A3B3CL24 E9HN60 K7J219 A0A2U9CEW3 Q5ZK92-2 A0A3B4ZRW5 A0A146VQQ2 I3JUV2
A0A2W1B7L8 A0A1Y1KFA1 A0A1Y1KL42 A0A158NSN8 A0A1Y1KHQ5 A0A1Y1KMR0 A0A1Q3EUM0 A0A1Q3EUZ1 D6WR93 N6SRM3 A0A3B0JKI5 A0A034W3J6 A0A1W4U7W3 A0A0K8UYH1 A0A0R3NH26 A0A0A1WSU7 A0A1L8DU08 A0A1L8DTP0 A0A1I8NNF4 E0VYI4 A0A0M4F6S0 W8C8D8 A0A0Q9WIN0 A0A1W4W830 A0A0P8XY81 A0A0Q9WTF8 A0A1B6I365 A0A0Q9WY76 A0A1W4ULF9 A0A126GV13 A0A0B4LHJ5 A0A146L6H0 A0A126GV10 A0A0A9ZEL2 A0A0A9ZIL4 B4HGG6 Q8I0P1 A0A1B6KIZ5 A0A088A016 B4QSF0 F4W8A4 E2AI24 A0A195CAZ2 A0A1B6GJT6 A0A069DV18 A0A0V0G8F5 A0A224XLX6 A0A0P4VTL2 A0A182P867 A0A1B0D797 A0A0P4W909 A0A3Q0JEE6 A0A2J7Q240 A0A1B6DYR9 R7TU42 A0A2R5L4N1 A0A0L7R7C7 E2BI57 A0A2A3E4B6 A0A3Q2E8P7 A0A3L8DA27 A0A026WUX2 A0A151I5Z7 A0A195DUQ1 A0A154P359 A0A1B6CUX1 A0A1Z5KY73 A0A2L2YK85 A0A0C9PQF2 A0A151XDP6 A0A0T6B521 A0A2J7Q222 A0A2J7Q234 A0A0C9QYQ7 A0A3P8UBX3 A0A3Q1CK62 A0A1B6G657 A0A146MUH2 A0A3Q2TUE9 A0A3B3YLW5 A0A3Q1EVP2 M3ZW57 A0A1L1RP85 A0A146RUZ8 A0A2M4BN30 A0A1V4JL95 A0A3B3CL24 E9HN60 K7J219 A0A2U9CEW3 Q5ZK92-2 A0A3B4ZRW5 A0A146VQQ2 I3JUV2
EC Number
5.6.1.1
Pubmed
26354079
19121390
22118469
28756777
28004739
21347285
+ More
18362917 19820115 23537049 25348373 15632085 17994087 25830018 20566863 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 25401762 12537569 12908108 15242610 15562320 15823537 16276413 17452528 18327897 19341724 23122959 25875445 18202664 21719571 20798317 26334808 27129103 23254933 30249741 24508170 28528879 26561354 23542700 15592404 29451363 21292972 20075255 15642098 25186727
18362917 19820115 23537049 25348373 15632085 17994087 25830018 20566863 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 25401762 12537569 12908108 15242610 15562320 15823537 16276413 17452528 18327897 19341724 23122959 25875445 18202664 21719571 20798317 26334808 27129103 23254933 30249741 24508170 28528879 26561354 23542700 15592404 29451363 21292972 20075255 15642098 25186727
EMBL
NWSH01007192
PCG63044.1
KQ459324
KPJ01492.1
PCG63046.1
KQ460973
+ More
KPJ10059.1 BABH01019775 BABH01019776 BABH01019777 BABH01019778 AGBW02011239 OWR46982.1 KZ150250 PZC71832.1 GEZM01085201 JAV60129.1 GEZM01085204 JAV60126.1 ADTU01025081 ADTU01025082 GEZM01085202 JAV60128.1 GEZM01085203 JAV60127.1 GFDL01016026 JAV19019.1 GFDL01016027 JAV19018.1 KQ971354 EFA07037.1 APGK01059436 APGK01059437 KB741293 KB632046 ENN70269.1 ERL88278.1 OUUW01000007 SPP82735.1 GAKP01009678 JAC49274.1 GDHF01020658 JAI31656.1 CM000070 KRT00353.1 GBXI01012712 JAD01580.1 GFDF01004204 JAV09880.1 GFDF01004265 JAV09819.1 DS235845 EEB18440.1 CP012526 ALC47445.1 GAMC01007521 GAMC01007519 JAB99034.1 CH940650 KRF83798.1 CH902617 KPU79694.1 CH964232 KRF99403.1 GECU01026344 JAS81362.1 CH933806 KRG00934.1 AE014297 ALI30635.1 AHN57493.1 GDHC01014581 JAQ04048.1 ALI30634.1 GBHO01000730 JAG42874.1 GBHO01000729 GBRD01006505 JAG42875.1 JAG59316.1 CH480815 AY069522 BT001254 BT001351 BT044258 GEBQ01028549 JAT11428.1 CM000364 GL887898 EGI69599.1 GL439621 EFN66932.1 KQ978023 KYM97977.1 GECZ01007062 JAS62707.1 GBGD01000996 JAC87893.1 GECL01001705 JAP04419.1 GFTR01006986 JAW09440.1 GDKW01003380 JAI53215.1 AJVK01026762 AJVK01026763 GDRN01107862 JAI57357.1 NEVH01019373 PNF22640.1 GEDC01006501 JAS30797.1 AMQN01012240 KB309387 ELT94540.1 GGLE01000281 MBY04407.1 KQ414643 KOC66661.1 GL448450 EFN84628.1 KZ288379 PBC26603.1 QOIP01000011 RLU16973.1 KK107111 EZA58904.1 KQ976400 KYM92670.1 KQ980322 KYN16603.1 KQ434809 KZC06365.1 GEDC01020127 JAS17171.1 GFJQ02007072 JAV99897.1 IAAA01030916 LAA08532.1 GBYB01003436 GBYB01003438 JAG73203.1 JAG73205.1 KQ982275 KYQ58440.1 LJIG01009785 KRT82385.1 PNF22641.1 PNF22642.1 GBYB01005787 JAG75554.1 GECZ01011849 JAS57920.1 GCES01163570 JAQ22752.1 AADN05000005 GCES01114120 JAQ72202.1 GGFJ01005346 MBW54487.1 LSYS01007194 OPJ72527.1 GL732693 EFX66831.1 CP026257 AWP14239.1 EU849599 EU849600 AJ720192 AADN03003400 AADN03003564 GCES01066581 JAR19742.1 AERX01006480
KPJ10059.1 BABH01019775 BABH01019776 BABH01019777 BABH01019778 AGBW02011239 OWR46982.1 KZ150250 PZC71832.1 GEZM01085201 JAV60129.1 GEZM01085204 JAV60126.1 ADTU01025081 ADTU01025082 GEZM01085202 JAV60128.1 GEZM01085203 JAV60127.1 GFDL01016026 JAV19019.1 GFDL01016027 JAV19018.1 KQ971354 EFA07037.1 APGK01059436 APGK01059437 KB741293 KB632046 ENN70269.1 ERL88278.1 OUUW01000007 SPP82735.1 GAKP01009678 JAC49274.1 GDHF01020658 JAI31656.1 CM000070 KRT00353.1 GBXI01012712 JAD01580.1 GFDF01004204 JAV09880.1 GFDF01004265 JAV09819.1 DS235845 EEB18440.1 CP012526 ALC47445.1 GAMC01007521 GAMC01007519 JAB99034.1 CH940650 KRF83798.1 CH902617 KPU79694.1 CH964232 KRF99403.1 GECU01026344 JAS81362.1 CH933806 KRG00934.1 AE014297 ALI30635.1 AHN57493.1 GDHC01014581 JAQ04048.1 ALI30634.1 GBHO01000730 JAG42874.1 GBHO01000729 GBRD01006505 JAG42875.1 JAG59316.1 CH480815 AY069522 BT001254 BT001351 BT044258 GEBQ01028549 JAT11428.1 CM000364 GL887898 EGI69599.1 GL439621 EFN66932.1 KQ978023 KYM97977.1 GECZ01007062 JAS62707.1 GBGD01000996 JAC87893.1 GECL01001705 JAP04419.1 GFTR01006986 JAW09440.1 GDKW01003380 JAI53215.1 AJVK01026762 AJVK01026763 GDRN01107862 JAI57357.1 NEVH01019373 PNF22640.1 GEDC01006501 JAS30797.1 AMQN01012240 KB309387 ELT94540.1 GGLE01000281 MBY04407.1 KQ414643 KOC66661.1 GL448450 EFN84628.1 KZ288379 PBC26603.1 QOIP01000011 RLU16973.1 KK107111 EZA58904.1 KQ976400 KYM92670.1 KQ980322 KYN16603.1 KQ434809 KZC06365.1 GEDC01020127 JAS17171.1 GFJQ02007072 JAV99897.1 IAAA01030916 LAA08532.1 GBYB01003436 GBYB01003438 JAG73203.1 JAG73205.1 KQ982275 KYQ58440.1 LJIG01009785 KRT82385.1 PNF22641.1 PNF22642.1 GBYB01005787 JAG75554.1 GECZ01011849 JAS57920.1 GCES01163570 JAQ22752.1 AADN05000005 GCES01114120 JAQ72202.1 GGFJ01005346 MBW54487.1 LSYS01007194 OPJ72527.1 GL732693 EFX66831.1 CP026257 AWP14239.1 EU849599 EU849600 AJ720192 AADN03003400 AADN03003564 GCES01066581 JAR19742.1 AERX01006480
Proteomes
UP000218220
UP000053268
UP000053240
UP000005204
UP000007151
UP000005205
+ More
UP000007266 UP000019118 UP000030742 UP000268350 UP000192221 UP000001819 UP000095300 UP000009046 UP000092553 UP000008792 UP000192223 UP000007801 UP000007798 UP000009192 UP000000803 UP000001292 UP000005203 UP000000304 UP000007755 UP000000311 UP000078542 UP000075885 UP000092462 UP000079169 UP000235965 UP000014760 UP000053825 UP000008237 UP000242457 UP000265020 UP000279307 UP000053097 UP000078540 UP000078492 UP000076502 UP000075809 UP000265080 UP000257160 UP000265000 UP000261480 UP000257200 UP000002852 UP000000539 UP000190648 UP000261560 UP000000305 UP000002358 UP000246464 UP000261400 UP000005207
UP000007266 UP000019118 UP000030742 UP000268350 UP000192221 UP000001819 UP000095300 UP000009046 UP000092553 UP000008792 UP000192223 UP000007801 UP000007798 UP000009192 UP000000803 UP000001292 UP000005203 UP000000304 UP000007755 UP000000311 UP000078542 UP000075885 UP000092462 UP000079169 UP000235965 UP000014760 UP000053825 UP000008237 UP000242457 UP000265020 UP000279307 UP000053097 UP000078540 UP000078492 UP000076502 UP000075809 UP000265080 UP000257160 UP000265000 UP000261480 UP000257200 UP000002852 UP000000539 UP000190648 UP000261560 UP000000305 UP000002358 UP000246464 UP000261400 UP000005207
Interpro
Gene 3D
ProteinModelPortal
A0A2A4IUU0
A0A194Q7H6
A0A2A4IU74
A0A0N1I5K3
H9J6D3
A0A212EZS7
+ More
A0A2W1B7L8 A0A1Y1KFA1 A0A1Y1KL42 A0A158NSN8 A0A1Y1KHQ5 A0A1Y1KMR0 A0A1Q3EUM0 A0A1Q3EUZ1 D6WR93 N6SRM3 A0A3B0JKI5 A0A034W3J6 A0A1W4U7W3 A0A0K8UYH1 A0A0R3NH26 A0A0A1WSU7 A0A1L8DU08 A0A1L8DTP0 A0A1I8NNF4 E0VYI4 A0A0M4F6S0 W8C8D8 A0A0Q9WIN0 A0A1W4W830 A0A0P8XY81 A0A0Q9WTF8 A0A1B6I365 A0A0Q9WY76 A0A1W4ULF9 A0A126GV13 A0A0B4LHJ5 A0A146L6H0 A0A126GV10 A0A0A9ZEL2 A0A0A9ZIL4 B4HGG6 Q8I0P1 A0A1B6KIZ5 A0A088A016 B4QSF0 F4W8A4 E2AI24 A0A195CAZ2 A0A1B6GJT6 A0A069DV18 A0A0V0G8F5 A0A224XLX6 A0A0P4VTL2 A0A182P867 A0A1B0D797 A0A0P4W909 A0A3Q0JEE6 A0A2J7Q240 A0A1B6DYR9 R7TU42 A0A2R5L4N1 A0A0L7R7C7 E2BI57 A0A2A3E4B6 A0A3Q2E8P7 A0A3L8DA27 A0A026WUX2 A0A151I5Z7 A0A195DUQ1 A0A154P359 A0A1B6CUX1 A0A1Z5KY73 A0A2L2YK85 A0A0C9PQF2 A0A151XDP6 A0A0T6B521 A0A2J7Q222 A0A2J7Q234 A0A0C9QYQ7 A0A3P8UBX3 A0A3Q1CK62 A0A1B6G657 A0A146MUH2 A0A3Q2TUE9 A0A3B3YLW5 A0A3Q1EVP2 M3ZW57 A0A1L1RP85 A0A146RUZ8 A0A2M4BN30 A0A1V4JL95 A0A3B3CL24 E9HN60 K7J219 A0A2U9CEW3 Q5ZK92-2 A0A3B4ZRW5 A0A146VQQ2 I3JUV2
A0A2W1B7L8 A0A1Y1KFA1 A0A1Y1KL42 A0A158NSN8 A0A1Y1KHQ5 A0A1Y1KMR0 A0A1Q3EUM0 A0A1Q3EUZ1 D6WR93 N6SRM3 A0A3B0JKI5 A0A034W3J6 A0A1W4U7W3 A0A0K8UYH1 A0A0R3NH26 A0A0A1WSU7 A0A1L8DU08 A0A1L8DTP0 A0A1I8NNF4 E0VYI4 A0A0M4F6S0 W8C8D8 A0A0Q9WIN0 A0A1W4W830 A0A0P8XY81 A0A0Q9WTF8 A0A1B6I365 A0A0Q9WY76 A0A1W4ULF9 A0A126GV13 A0A0B4LHJ5 A0A146L6H0 A0A126GV10 A0A0A9ZEL2 A0A0A9ZIL4 B4HGG6 Q8I0P1 A0A1B6KIZ5 A0A088A016 B4QSF0 F4W8A4 E2AI24 A0A195CAZ2 A0A1B6GJT6 A0A069DV18 A0A0V0G8F5 A0A224XLX6 A0A0P4VTL2 A0A182P867 A0A1B0D797 A0A0P4W909 A0A3Q0JEE6 A0A2J7Q240 A0A1B6DYR9 R7TU42 A0A2R5L4N1 A0A0L7R7C7 E2BI57 A0A2A3E4B6 A0A3Q2E8P7 A0A3L8DA27 A0A026WUX2 A0A151I5Z7 A0A195DUQ1 A0A154P359 A0A1B6CUX1 A0A1Z5KY73 A0A2L2YK85 A0A0C9PQF2 A0A151XDP6 A0A0T6B521 A0A2J7Q222 A0A2J7Q234 A0A0C9QYQ7 A0A3P8UBX3 A0A3Q1CK62 A0A1B6G657 A0A146MUH2 A0A3Q2TUE9 A0A3B3YLW5 A0A3Q1EVP2 M3ZW57 A0A1L1RP85 A0A146RUZ8 A0A2M4BN30 A0A1V4JL95 A0A3B3CL24 E9HN60 K7J219 A0A2U9CEW3 Q5ZK92-2 A0A3B4ZRW5 A0A146VQQ2 I3JUV2
PDB
6NYV
E-value=7.87603e-115,
Score=1059
Ontologies
GO
GO:0005813
GO:0008568
GO:0005874
GO:0008017
GO:0005524
GO:0031117
GO:0016021
GO:0005819
GO:0005737
GO:0034214
GO:0016853
GO:0031122
GO:0005634
GO:0016887
GO:0005694
GO:0005811
GO:0050803
GO:0000022
GO:0008344
GO:0051301
GO:0030154
GO:0007399
GO:0007079
GO:0000502
GO:0045886
GO:2000331
GO:0035099
GO:0045887
GO:0045834
GO:0048691
GO:0051013
GO:0031594
GO:0008021
GO:1900074
GO:0050775
GO:0043195
GO:1900075
GO:0019226
GO:0043014
GO:0007626
GO:0015630
GO:0048167
GO:0000226
GO:0000070
GO:0006888
GO:0007409
GO:0048471
GO:0032506
GO:0007032
GO:0072593
GO:0045773
GO:0048675
GO:0043066
GO:0021955
GO:0031965
GO:0005768
GO:0051228
GO:0010458
GO:0051260
GO:0031468
GO:0005654
GO:0001578
GO:0005829
GO:0030496
GO:0019896
GO:1904115
GO:0000281
GO:0005783
GO:0048487
GO:0090148
GO:0032467
GO:0008089
GO:0006436
GO:0004812
GO:0006418
GO:0004222
GO:0031012
GO:0008233
Topology
Subcellular location
Membrane
Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Cytoplasm Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Cytoskeleton Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Microtubule organizing center Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Centrosome Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Chromosome Forms an intramembrane hairpin-like structure in the membrane. Colocalizes with cellular microtubule arrays. Localizes to chromosomes from prometaphase/metaphase to anaphase, and this requires microtubules. Localizes to discrete punctate cytoplasmic foci which may correspond to secretory vesicles. With evidence from 1 publications.
Lipid droplet Forms an intramembrane hairpin-like structure in the membrane. Colocalizes with cellular microtubule arrays. Localizes to chromosomes from prometaphase/metaphase to anaphase, and this requires microtubules. Localizes to discrete punctate cytoplasmic foci which may correspond to secretory vesicles. With evidence from 1 publications.
Perinuclear region Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Nucleus Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Cytoplasm Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Cytoskeleton Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Microtubule organizing center Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Centrosome Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Chromosome Forms an intramembrane hairpin-like structure in the membrane. Colocalizes with cellular microtubule arrays. Localizes to chromosomes from prometaphase/metaphase to anaphase, and this requires microtubules. Localizes to discrete punctate cytoplasmic foci which may correspond to secretory vesicles. With evidence from 1 publications.
Lipid droplet Forms an intramembrane hairpin-like structure in the membrane. Colocalizes with cellular microtubule arrays. Localizes to chromosomes from prometaphase/metaphase to anaphase, and this requires microtubules. Localizes to discrete punctate cytoplasmic foci which may correspond to secretory vesicles. With evidence from 1 publications.
Perinuclear region Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
Nucleus Forms an intramembrane hairpin-like structure in the membrane. With evidence from 1 publications.
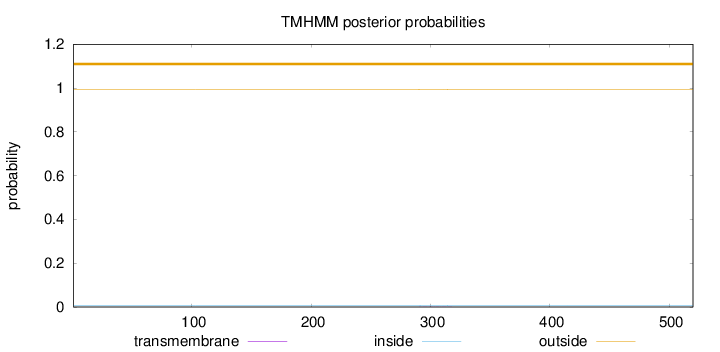
Length:
520
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01278
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00613
outside
1 - 520
Population Genetic Test Statistics
Pi
269.31313
Theta
195.190418
Tajima's D
0.949433
CLR
0.389685
CSRT
0.649067546622669
Interpretation
Uncertain
