Gene
KWMTBOMO15335
Pre Gene Modal
BGIBMGA005070
Annotation
PREDICTED:_dolichyl-phosphate-mannose--protein_mannosyltransferase_4_isoform_X2_[Amyelois_transitella]
Full name
Protein O-mannosyl-transferase 2
Alternative Name
Dolichyl-phosphate-mannose--protein mannosyltransferase 2
Protein twisted
Protein twisted
Location in the cell
PlasmaMembrane Reliability : 3.962
Sequence
CDS
ATGGCAAGTTGGTACATAAACAGGACCTTCTTCTTTGATGTACATCCACCACTTGGCAAGATGCTAATAGCATTGTCTGGCAAACTATCAGGGTATGATGGTACATTTCCATTTGAGAAACCTGGAGATAAGTATGATGGTGCGAGGTATCAGGGTATGAGACTATTTTGTACAACGTTAGGTGCTTTGGTTATTCCCCTAACCTTTCTGTCGATATGGGAGATGACTAAATCATTGGAAGCCACGTTCATAGGAACGTTGCTCTTGTTGTGTGATGTTGGCTTTATGACTTTAAACAGATACATCTTATTAGATCCAATCTTGCTGTTCTTCATGAGTTGTTCAATATATGGATGCTTTAAGGTTTCTACTTTAACCAATGACGGCTTACTCCCTTTTTCTCCGAGGATGCTGTCATGGCAAATTTTCTTGGGTACATCATTAGCTTGCACGATGTCCGTCAAGTTTGTGGGATTGTTCGTTGTTCTATTTGTTGGTCTACGGACAGTTGCGGACTTATGGAATATTCTGGGTGATTTATCGAAACCTGTTAAATTCACCATCACACATCTCATTGTCAGAACATTAACCCTCATTGTGTGGCCATTCGTTCTGTACGTGTTCTTCTTTTGGATACATCTCACGGTACTGAGTAAAAGCGGCAACGGTGACGGTTTCTACTCGTCCGGGTTCCAAGCACGTCTCGAAGGGAACAGCCTGCACAACGCCAGCGCGCCACGGGTGCTCGCTTACGGAGCTCTCGTCACTTTGAAGAATCATAGAACTGGCGGGGGGTACTTGCACTCACACCACCACTTGTACCCAGCGGGGGTGGGAGCGAGACAGCAACAGATAACAACATACACTCACAAAGATGAGAATAATCGTTGGCAAATAAAACCGTACAACCGCGAACGAGTCGAGGGGACGGTGGTGGTGCGCAGCGGCGATCTGGTGCGCGTCACGCACGCGCCCACGGGCCGCAACCTGCATTCGCACCGCGAGCGAGCGCCTCTCACCGCCAAGCACCTGCAGGTCACCGGCTACGGGGAGGACGGCGTAGGTGACGCCAACGACGTGTGGAAGATTGTCATATCGGGCGGTAAAGATGGCGAAGAGATCCAAACTGTTCGAAGCAAACTTATGCTTGTTCATTATTTGCAGGCGTGTGTATTGACGACAACAGGGAAGCAACTACCTAAATGGGGCTACGAACAGCAGGAAGTCTCCTGCAATCCCAATTTAAGAGACAAGAACGCGCTGTGGAACATAGAGGATAACGTTTTTGATGATCTGCCTAAAGTGAGTTTCGAATCTTACGCCTCTGGTTTCCTCGAACGTTTCTACGAATCGCACGCCGTCATGTTCCAGGGGAATTCCGGACTCAAACCGAAGGAAGGGGAAGTCACCTCGCAGCCGTGGCAATGGCCCATAAATTACAGGGGTCAATTTTTTTCGGGCAGCACTCACCGCATCTATTTATTAGGAAATCCAGTAGTATGGTGGACGAATCTTGCCTTTCTAATAATATTCTTCTTGGTGTATATCATCAACTCCATAAAAGAAAAGAGGGCAGAAGCTTTTGGTGTTTCTAGACAAGTTGGCGAAGGAAGTTTACTATTGAACGCAGCCGGCTGGACGTTCATAGGATGGGCTCTGCACTACATACCTTTCTGGGCTATGGGCAGAGTTTTGTATTTTCACCATTACTTTCCTGCCCTAGTCTTCAGCTCTATGCTTACAGGTATCTTAACTGAATATCTTCTAAGTAGTGTGCGGAGTTACTTGAGTCCGGAACTGGGAAAGACTGCGTATCATTGCATCATAGGCGTTGTAATATCAACAACAGTGTACAGTTTCTATTTGTTTTCACCACTAGCTTACGGTATGAGCGGACCGTTGGCTAGTGAACCGAATTCGACAATGGCAGGCCTTAAATGGCTTGAATCTTGGGAATTTTAA
Protein
MASWYINRTFFFDVHPPLGKMLIALSGKLSGYDGTFPFEKPGDKYDGARYQGMRLFCTTLGALVIPLTFLSIWEMTKSLEATFIGTLLLLCDVGFMTLNRYILLDPILLFFMSCSIYGCFKVSTLTNDGLLPFSPRMLSWQIFLGTSLACTMSVKFVGLFVVLFVGLRTVADLWNILGDLSKPVKFTITHLIVRTLTLIVWPFVLYVFFFWIHLTVLSKSGNGDGFYSSGFQARLEGNSLHNASAPRVLAYGALVTLKNHRTGGGYLHSHHHLYPAGVGARQQQITTYTHKDENNRWQIKPYNRERVEGTVVVRSGDLVRVTHAPTGRNLHSHRERAPLTAKHLQVTGYGEDGVGDANDVWKIVISGGKDGEEIQTVRSKLMLVHYLQACVLTTTGKQLPKWGYEQQEVSCNPNLRDKNALWNIEDNVFDDLPKVSFESYASGFLERFYESHAVMFQGNSGLKPKEGEVTSQPWQWPINYRGQFFSGSTHRIYLLGNPVVWWTNLAFLIIFFLVYIINSIKEKRAEAFGVSRQVGEGSLLLNAAGWTFIGWALHYIPFWAMGRVLYFHHYFPALVFSSMLTGILTEYLLSSVRSYLSPELGKTAYHCIIGVVISTTVYSFYLFSPLAYGMSGPLASEPNSTMAGLKWLESWEF
Summary
Description
Rt/POMT1 and tw/POMT2 function as a protein O-mannosyltransferase in association with each other to generate and maintain normal muscle development.
Catalytic Activity
dolichyl beta-D-mannosyl phosphate + L-seryl-[protein] = 3-O-(alpha-D-mannosyl)-L-seryl-[protein] + dolichyl phosphate + H(+)
dolichyl beta-D-mannosyl phosphate + L-threonyl-[protein] = 3-O-(alpha-D-mannosyl)-L-threonyl-[protein] + dolichyl phosphate + H(+)
dolichyl beta-D-mannosyl phosphate + L-threonyl-[protein] = 3-O-(alpha-D-mannosyl)-L-threonyl-[protein] + dolichyl phosphate + H(+)
Subunit
Interacts with Rt/POMT1.
Similarity
Belongs to the glycosyltransferase 39 family.
Keywords
Complete proteome
Developmental protein
Endoplasmic reticulum
Glycoprotein
Glycosyltransferase
Membrane
Reference proteome
Repeat
Transferase
Transmembrane
Transmembrane helix
Polymorphism
Feature
chain Protein O-mannosyl-transferase 2
Uniprot
A0A2A4K8F2
A0A2H1VRD3
A0A3S2MAM0
A0A194Q950
A0A212EZP6
A0A2W1BP81
+ More
A0A1B6CJ65 A0A2J7RCJ0 J3JYN2 A0A067QRN3 A0A1Y1KV40 A0A0L0BV96 A0A1B6JB66 N6T575 U4UD25 A0A1B0GJC6 B4JWZ9 A0A1S4FZS8 Q16IB3 B4MT03 K7IQD7 A0A084W6Z5 A0A182QTA3 A0A2S2QAA7 A0A232F425 A0A182HSB3 A0A195C6M6 A0A182JSV5 A0A182XAH3 A0A151XBR6 F4WG18 A0A151JWJ4 A0A0K8VQT0 Q7PGK7 E2AI29 A0A182Y6R6 A0A182VG21 Q29IL2 B4H2N6 B4L836 A0A182IU27 A0A1J1HLJ6 A0A0K8VJP6 M9PG53 Q9W5D4 A0A3L8D7U2 A0A1Y1L2D7 B4M2Y4 B4PX34 B3P9A1 A0A1W4VFF0 A0A3B0JYH9 A0A0A1X547 B4R7H9 A0A158NTG6 W8BFB3 A0A2S2PQ03 B3MR19 A0A195BG62 A0A0L7QTS8 A0A026WBI2 A0A088AB89 A0A0P9A9N4 A0A2A3ERK7 A0A224XCI9 A0A069DVR1 A0A0A9W580 A0A023F4R5 A0A0K8UDX0 T1J5X4 A0A1E1XP87 A0A1Y1KXH3 A0A1D2NH85 A0A336M336 A0A1I8NX85 A0A131XHU0 A0A1E1X910 A0A182G599 A0A2R5LEW3 A0A131YIH4 A0A182LE96 A0A224Y8Z4 A0A0M8ZU82 A0A182FQ56 A0A1B6FFG6 A0A0P5QAM8 A0A182NR86 A0A182VZX6 T1KER1 L7ME23 A0A154P310 L7LTG1 A0A0P6FZX0 A0A0N8D032 A0A0P5DI89 A0A0P6D3K9 A0A0P5NDQ4 A0A0P5YA67 A0A0P6D3L4 A0A0P6EDS0
A0A1B6CJ65 A0A2J7RCJ0 J3JYN2 A0A067QRN3 A0A1Y1KV40 A0A0L0BV96 A0A1B6JB66 N6T575 U4UD25 A0A1B0GJC6 B4JWZ9 A0A1S4FZS8 Q16IB3 B4MT03 K7IQD7 A0A084W6Z5 A0A182QTA3 A0A2S2QAA7 A0A232F425 A0A182HSB3 A0A195C6M6 A0A182JSV5 A0A182XAH3 A0A151XBR6 F4WG18 A0A151JWJ4 A0A0K8VQT0 Q7PGK7 E2AI29 A0A182Y6R6 A0A182VG21 Q29IL2 B4H2N6 B4L836 A0A182IU27 A0A1J1HLJ6 A0A0K8VJP6 M9PG53 Q9W5D4 A0A3L8D7U2 A0A1Y1L2D7 B4M2Y4 B4PX34 B3P9A1 A0A1W4VFF0 A0A3B0JYH9 A0A0A1X547 B4R7H9 A0A158NTG6 W8BFB3 A0A2S2PQ03 B3MR19 A0A195BG62 A0A0L7QTS8 A0A026WBI2 A0A088AB89 A0A0P9A9N4 A0A2A3ERK7 A0A224XCI9 A0A069DVR1 A0A0A9W580 A0A023F4R5 A0A0K8UDX0 T1J5X4 A0A1E1XP87 A0A1Y1KXH3 A0A1D2NH85 A0A336M336 A0A1I8NX85 A0A131XHU0 A0A1E1X910 A0A182G599 A0A2R5LEW3 A0A131YIH4 A0A182LE96 A0A224Y8Z4 A0A0M8ZU82 A0A182FQ56 A0A1B6FFG6 A0A0P5QAM8 A0A182NR86 A0A182VZX6 T1KER1 L7ME23 A0A154P310 L7LTG1 A0A0P6FZX0 A0A0N8D032 A0A0P5DI89 A0A0P6D3K9 A0A0P5NDQ4 A0A0P5YA67 A0A0P6D3L4 A0A0P6EDS0
EC Number
2.4.1.109
Pubmed
26354079
22118469
28756777
22516182
24845553
28004739
+ More
26108605 23537049 17994087 17510324 20075255 24438588 28648823 21719571 12364791 14747013 17210077 20798317 25244985 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15271988 16219785 10731137 12537569 30249741 17550304 25830018 21347285 24495485 24508170 26334808 25401762 25474469 29209593 27289101 28049606 28503490 26483478 26830274 20966253 28797301 25576852
26108605 23537049 17994087 17510324 20075255 24438588 28648823 21719571 12364791 14747013 17210077 20798317 25244985 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15271988 16219785 10731137 12537569 30249741 17550304 25830018 21347285 24495485 24508170 26334808 25401762 25474469 29209593 27289101 28049606 28503490 26483478 26830274 20966253 28797301 25576852
EMBL
NWSH01000029
PCG80555.1
ODYU01003989
SOQ43411.1
RSAL01000001
RVE55240.1
+ More
KQ459324 KPJ01500.1 AGBW02011239 OWR46968.1 KZ150042 PZC74550.1 GEDC01023762 JAS13536.1 NEVH01005885 PNF38553.1 BT128361 AEE63319.1 KK853897 KDQ71515.1 GEZM01073205 JAV65279.1 JRES01001281 KNC23957.1 GECU01011307 JAS96399.1 APGK01052268 KB741211 ENN72818.1 KB632263 ERL90952.1 AJWK01020147 AJWK01020148 AJWK01020149 CH916376 EDV95275.1 CH478090 EAT34004.1 CH963851 EDW75242.2 ATLV01021070 KE525312 KFB45989.1 AXCN02001468 GGMS01005470 MBY74673.1 NNAY01001084 OXU25203.1 APCN01000260 KQ978205 KYM96514.1 KQ982320 KYQ57789.1 GL888128 EGI66785.1 KQ981639 KYN38691.1 GDHF01011092 JAI41222.1 AAAB01008859 EAA44894.5 GL439621 EFN66937.1 CH379063 EAL32641.1 CH479204 EDW30603.1 CH933814 EDW05611.1 CVRI01000010 CRK88936.1 GDHF01013221 JAI39093.1 AE014298 AGB94948.1 AB176551 AL031583 AY095525 AAF45548.1 AAM12256.1 BAD54755.1 CAA20897.1 QOIP01000012 RLU16152.1 GEZM01073206 JAV65277.1 CH940651 EDW65159.2 CM000162 EDX00820.1 CH954183 EDV45397.1 OUUW01000003 SPP78436.1 GBXI01007843 JAD06449.1 CM000366 EDX16790.1 ADTU01025893 GAMC01006575 JAB99980.1 GGMR01018759 MBY31378.1 CH902622 EDV34224.1 KQ976488 KYM83582.1 KQ414741 KOC61959.1 KK107341 EZA52404.1 KPU75029.1 KZ288194 PBC33846.1 GFTR01007742 JAW08684.1 GBGD01000701 JAC88188.1 GBHO01043580 GBHO01043579 GBHO01032657 GBHO01032655 GBHO01032654 GBHO01032650 JAG00024.1 JAG00025.1 JAG10947.1 JAG10949.1 JAG10950.1 JAG10954.1 GBBI01002743 JAC15969.1 GDHF01027729 JAI24585.1 JH431868 GFAA01002297 JAU01138.1 GEZM01073212 JAV65268.1 LJIJ01000040 ODN04599.1 UFQT01000480 SSX24662.1 GEFH01003360 JAP65221.1 GFAC01003657 JAT95531.1 JXUM01043074 JXUM01043075 JXUM01043076 JXUM01043077 KQ561349 KXJ78868.1 GGLE01003910 MBY08036.1 GEDV01009473 JAP79084.1 GFPF01000893 MAA12039.1 KQ435848 KOX71057.1 GECZ01020828 JAS48941.1 GDIQ01123522 JAL28204.1 CAEY01000028 GACK01002799 JAA62235.1 KQ434809 KZC06223.1 GACK01010137 JAA54897.1 GDIQ01058599 JAN36138.1 GDIP01081991 JAM21724.1 GDIP01157080 JAJ66322.1 GDIQ01083289 JAN11448.1 GDIQ01146776 JAL04950.1 GDIP01254308 GDIP01217924 GDIP01199477 GDIP01097424 GDIP01093285 GDIP01060841 GDIP01060840 GDIP01058659 JAM42874.1 GDIQ01125425 GDIQ01110433 GDIQ01088186 GDIQ01083288 GDIQ01046510 GDIQ01042209 GDIQ01039406 JAN11449.1 GDIQ01064247 GDIQ01064246 JAN30490.1
KQ459324 KPJ01500.1 AGBW02011239 OWR46968.1 KZ150042 PZC74550.1 GEDC01023762 JAS13536.1 NEVH01005885 PNF38553.1 BT128361 AEE63319.1 KK853897 KDQ71515.1 GEZM01073205 JAV65279.1 JRES01001281 KNC23957.1 GECU01011307 JAS96399.1 APGK01052268 KB741211 ENN72818.1 KB632263 ERL90952.1 AJWK01020147 AJWK01020148 AJWK01020149 CH916376 EDV95275.1 CH478090 EAT34004.1 CH963851 EDW75242.2 ATLV01021070 KE525312 KFB45989.1 AXCN02001468 GGMS01005470 MBY74673.1 NNAY01001084 OXU25203.1 APCN01000260 KQ978205 KYM96514.1 KQ982320 KYQ57789.1 GL888128 EGI66785.1 KQ981639 KYN38691.1 GDHF01011092 JAI41222.1 AAAB01008859 EAA44894.5 GL439621 EFN66937.1 CH379063 EAL32641.1 CH479204 EDW30603.1 CH933814 EDW05611.1 CVRI01000010 CRK88936.1 GDHF01013221 JAI39093.1 AE014298 AGB94948.1 AB176551 AL031583 AY095525 AAF45548.1 AAM12256.1 BAD54755.1 CAA20897.1 QOIP01000012 RLU16152.1 GEZM01073206 JAV65277.1 CH940651 EDW65159.2 CM000162 EDX00820.1 CH954183 EDV45397.1 OUUW01000003 SPP78436.1 GBXI01007843 JAD06449.1 CM000366 EDX16790.1 ADTU01025893 GAMC01006575 JAB99980.1 GGMR01018759 MBY31378.1 CH902622 EDV34224.1 KQ976488 KYM83582.1 KQ414741 KOC61959.1 KK107341 EZA52404.1 KPU75029.1 KZ288194 PBC33846.1 GFTR01007742 JAW08684.1 GBGD01000701 JAC88188.1 GBHO01043580 GBHO01043579 GBHO01032657 GBHO01032655 GBHO01032654 GBHO01032650 JAG00024.1 JAG00025.1 JAG10947.1 JAG10949.1 JAG10950.1 JAG10954.1 GBBI01002743 JAC15969.1 GDHF01027729 JAI24585.1 JH431868 GFAA01002297 JAU01138.1 GEZM01073212 JAV65268.1 LJIJ01000040 ODN04599.1 UFQT01000480 SSX24662.1 GEFH01003360 JAP65221.1 GFAC01003657 JAT95531.1 JXUM01043074 JXUM01043075 JXUM01043076 JXUM01043077 KQ561349 KXJ78868.1 GGLE01003910 MBY08036.1 GEDV01009473 JAP79084.1 GFPF01000893 MAA12039.1 KQ435848 KOX71057.1 GECZ01020828 JAS48941.1 GDIQ01123522 JAL28204.1 CAEY01000028 GACK01002799 JAA62235.1 KQ434809 KZC06223.1 GACK01010137 JAA54897.1 GDIQ01058599 JAN36138.1 GDIP01081991 JAM21724.1 GDIP01157080 JAJ66322.1 GDIQ01083289 JAN11448.1 GDIQ01146776 JAL04950.1 GDIP01254308 GDIP01217924 GDIP01199477 GDIP01097424 GDIP01093285 GDIP01060841 GDIP01060840 GDIP01058659 JAM42874.1 GDIQ01125425 GDIQ01110433 GDIQ01088186 GDIQ01083288 GDIQ01046510 GDIQ01042209 GDIQ01039406 JAN11449.1 GDIQ01064247 GDIQ01064246 JAN30490.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000235965
UP000027135
+ More
UP000037069 UP000019118 UP000030742 UP000092461 UP000001070 UP000008820 UP000007798 UP000002358 UP000030765 UP000075886 UP000215335 UP000075840 UP000078542 UP000075881 UP000076407 UP000075809 UP000007755 UP000078541 UP000007062 UP000000311 UP000076408 UP000075903 UP000001819 UP000008744 UP000009192 UP000075880 UP000183832 UP000000803 UP000279307 UP000008792 UP000002282 UP000008711 UP000192221 UP000268350 UP000000304 UP000005205 UP000007801 UP000078540 UP000053825 UP000053097 UP000005203 UP000242457 UP000094527 UP000095300 UP000069940 UP000249989 UP000075882 UP000053105 UP000069272 UP000075884 UP000075920 UP000015104 UP000076502
UP000037069 UP000019118 UP000030742 UP000092461 UP000001070 UP000008820 UP000007798 UP000002358 UP000030765 UP000075886 UP000215335 UP000075840 UP000078542 UP000075881 UP000076407 UP000075809 UP000007755 UP000078541 UP000007062 UP000000311 UP000076408 UP000075903 UP000001819 UP000008744 UP000009192 UP000075880 UP000183832 UP000000803 UP000279307 UP000008792 UP000002282 UP000008711 UP000192221 UP000268350 UP000000304 UP000005205 UP000007801 UP000078540 UP000053825 UP000053097 UP000005203 UP000242457 UP000094527 UP000095300 UP000069940 UP000249989 UP000075882 UP000053105 UP000069272 UP000075884 UP000075920 UP000015104 UP000076502
Interpro
SUPFAM
SSF82109
SSF82109
ProteinModelPortal
A0A2A4K8F2
A0A2H1VRD3
A0A3S2MAM0
A0A194Q950
A0A212EZP6
A0A2W1BP81
+ More
A0A1B6CJ65 A0A2J7RCJ0 J3JYN2 A0A067QRN3 A0A1Y1KV40 A0A0L0BV96 A0A1B6JB66 N6T575 U4UD25 A0A1B0GJC6 B4JWZ9 A0A1S4FZS8 Q16IB3 B4MT03 K7IQD7 A0A084W6Z5 A0A182QTA3 A0A2S2QAA7 A0A232F425 A0A182HSB3 A0A195C6M6 A0A182JSV5 A0A182XAH3 A0A151XBR6 F4WG18 A0A151JWJ4 A0A0K8VQT0 Q7PGK7 E2AI29 A0A182Y6R6 A0A182VG21 Q29IL2 B4H2N6 B4L836 A0A182IU27 A0A1J1HLJ6 A0A0K8VJP6 M9PG53 Q9W5D4 A0A3L8D7U2 A0A1Y1L2D7 B4M2Y4 B4PX34 B3P9A1 A0A1W4VFF0 A0A3B0JYH9 A0A0A1X547 B4R7H9 A0A158NTG6 W8BFB3 A0A2S2PQ03 B3MR19 A0A195BG62 A0A0L7QTS8 A0A026WBI2 A0A088AB89 A0A0P9A9N4 A0A2A3ERK7 A0A224XCI9 A0A069DVR1 A0A0A9W580 A0A023F4R5 A0A0K8UDX0 T1J5X4 A0A1E1XP87 A0A1Y1KXH3 A0A1D2NH85 A0A336M336 A0A1I8NX85 A0A131XHU0 A0A1E1X910 A0A182G599 A0A2R5LEW3 A0A131YIH4 A0A182LE96 A0A224Y8Z4 A0A0M8ZU82 A0A182FQ56 A0A1B6FFG6 A0A0P5QAM8 A0A182NR86 A0A182VZX6 T1KER1 L7ME23 A0A154P310 L7LTG1 A0A0P6FZX0 A0A0N8D032 A0A0P5DI89 A0A0P6D3K9 A0A0P5NDQ4 A0A0P5YA67 A0A0P6D3L4 A0A0P6EDS0
A0A1B6CJ65 A0A2J7RCJ0 J3JYN2 A0A067QRN3 A0A1Y1KV40 A0A0L0BV96 A0A1B6JB66 N6T575 U4UD25 A0A1B0GJC6 B4JWZ9 A0A1S4FZS8 Q16IB3 B4MT03 K7IQD7 A0A084W6Z5 A0A182QTA3 A0A2S2QAA7 A0A232F425 A0A182HSB3 A0A195C6M6 A0A182JSV5 A0A182XAH3 A0A151XBR6 F4WG18 A0A151JWJ4 A0A0K8VQT0 Q7PGK7 E2AI29 A0A182Y6R6 A0A182VG21 Q29IL2 B4H2N6 B4L836 A0A182IU27 A0A1J1HLJ6 A0A0K8VJP6 M9PG53 Q9W5D4 A0A3L8D7U2 A0A1Y1L2D7 B4M2Y4 B4PX34 B3P9A1 A0A1W4VFF0 A0A3B0JYH9 A0A0A1X547 B4R7H9 A0A158NTG6 W8BFB3 A0A2S2PQ03 B3MR19 A0A195BG62 A0A0L7QTS8 A0A026WBI2 A0A088AB89 A0A0P9A9N4 A0A2A3ERK7 A0A224XCI9 A0A069DVR1 A0A0A9W580 A0A023F4R5 A0A0K8UDX0 T1J5X4 A0A1E1XP87 A0A1Y1KXH3 A0A1D2NH85 A0A336M336 A0A1I8NX85 A0A131XHU0 A0A1E1X910 A0A182G599 A0A2R5LEW3 A0A131YIH4 A0A182LE96 A0A224Y8Z4 A0A0M8ZU82 A0A182FQ56 A0A1B6FFG6 A0A0P5QAM8 A0A182NR86 A0A182VZX6 T1KER1 L7ME23 A0A154P310 L7LTG1 A0A0P6FZX0 A0A0N8D032 A0A0P5DI89 A0A0P6D3K9 A0A0P5NDQ4 A0A0P5YA67 A0A0P6D3L4 A0A0P6EDS0
PDB
1T9F
E-value=2.25752e-13,
Score=185
Ontologies
PATHWAY
GO
PANTHER
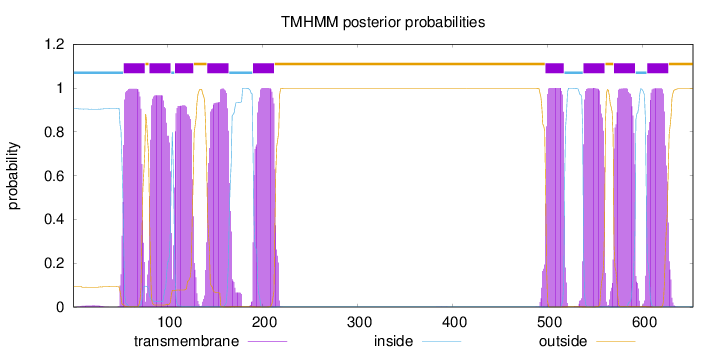
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
653
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
193.98175
Exp number, first 60 AAs:
7.83839
Total prob of N-in:
0.90631
inside
1 - 53
TMhelix
54 - 76
outside
77 - 80
TMhelix
81 - 103
inside
104 - 107
TMhelix
108 - 127
outside
128 - 141
TMhelix
142 - 164
inside
165 - 189
TMhelix
190 - 212
outside
213 - 497
TMhelix
498 - 517
inside
518 - 537
TMhelix
538 - 560
outside
561 - 569
TMhelix
570 - 592
inside
593 - 604
TMhelix
605 - 627
outside
628 - 653
Population Genetic Test Statistics
Pi
177.995606
Theta
151.482762
Tajima's D
1.032922
CLR
0.176536
CSRT
0.670066496675166
Interpretation
Uncertain
