Gene
KWMTBOMO15328 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005068
Annotation
PREDICTED:_neutral_and_basic_amino_acid_transport_protein_rBAT_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.043
Sequence
CDS
ATGTCGGAGCCACGGAAAAACCGTCTCTCGATTGATGGAGGGCAAGTGAAAGAGGATGAACACGTTGCAACCTACAAGGCTATACCAGAATCTGATACAGAATTCCGGTCGTCCAAAACGAACCTCGGCAAATCGAAGGAGAAGATATCTGCCGACGGCGCCGAAGAGAAACTATTGAAGAAGGAAGACGAGGCCAAAATCATAACCCGGGTGGACATGTCCGATGCGAAATATGTCGTCGGGGACCACAGGAATGGTGACGCGAAGATTGAACTCGATGCTAATAAACGGCAATTCACTGGGATGACGAGAGAGGAGGTACTGAAGTATGCTGACGATCCCTTCTGGGTGAACTTGCGTTGGTCGCTGTTCGTGTTGTTCTGGGTGGCCTGGCTGTGCATGCTCGCCGGGGCTATTGCAGTCATTGTACGCGCACCCAAGTGCGGCCCGCCCGAACCCAGGACATGGTACGAACTCGGTCCCCTGGTCGGACTGGACTTGGTCGATGCCGTCGAACCACAAGATTTACAACAGGATTTGGAACTGCTGAAGACATTCAAAGTTCAGGGCGTGTTCGTCCAAGTGCCGACTTACGAAGTGCTCGACAAACGTGACACACTGGACGAATTCAAAATTCTGTTTAACAAAATTAAGAAGATCGGAATAAGAGTGATTGTTGATTTGATACCGAACTACGTGTTCACCAACCATACCTGGTTCGTACAGAGTGAGAACAGCACTGAGCCCTATACTGATTATTTCATCTGGACCAAGGAACAACCTGCTAATTGGGTTTCAAAAATCAACATGCCAGCGTTTACACGAAGCGAGAAACGTCAGATGTTCTACCTGCATCAGTTCTGTGACAACTGCGCTGATCTTAATTTCGATAATCCTAAAGTTGTTGAGAAGTTCGACATGGTCCTGAAGGCCTGGATGGGTGCCGGCGCTAGTGGAGTCAGATTGAATAACGCGCGTCACTTGCTCGTCGAATTGCTTGAGGAGAAGACGCGCGTCGGTCGCGGCAGTAGCGTGGACGCGGATCACATGCGGTACGACTTCTGGGAGCACAAGCACACCACCGACCTGCCGAAGCTCAAGGAGCTCTTGGCTCGCTGGTCGAAGATCGTCACCAAAGAATCAGAGCCGACGGTGTTCACGCTGAAGGAAGATGGCTCCCTGCCGGACCTGATACTGCTGAACCACAACGTGTCGATGCTGCGGCCGCCGTCCGCCGCGCCCGTGCCCGTCACGGAGGACGCAGACGCGCTCGCCTCTAGAATAAACAAAACTCTTGGCTCTTGGCCTATTCTTCAGTTGACGGATACGAATGACGATCGTGAATTAGCATCGTTTGCTATGCTTCTACCGGCAGCACCTTTGTTGAATATAAAACAACTTTCAGCTGACGAAAACGACACCTCTCTTTTAGATCTGGTGAACCTCCGCAGCGACGCGTCGTTGTCGCACGGCGAGCACCACGTGGCGGCCGCGCCCGCGCGTAACGCAAGCGAGAACGTACTCATCGTTTGTGCAAGATGGAAAGAGGGTCACACCGGTTACATGGCGGTGTACAACCCGTCCCGGCAGGATCTCCACGCGAATCTCACTGCTGTACCCTCCGTGCCCGGGACGATAACAATTCAAAACGTCTCGCCAGCCGTCAAACTTGTCACCAACTATACGAAGAACTATCAGCCGGCAGAGGACGTGCTGGTCCCTGCTAAGTCCACAGTCGTTGTGTCTTATGTGCCTATAAAACATGAGAAGCGTGAAAAAGACGAATAA
Protein
MSEPRKNRLSIDGGQVKEDEHVATYKAIPESDTEFRSSKTNLGKSKEKISADGAEEKLLKKEDEAKIITRVDMSDAKYVVGDHRNGDAKIELDANKRQFTGMTREEVLKYADDPFWVNLRWSLFVLFWVAWLCMLAGAIAVIVRAPKCGPPEPRTWYELGPLVGLDLVDAVEPQDLQQDLELLKTFKVQGVFVQVPTYEVLDKRDTLDEFKILFNKIKKIGIRVIVDLIPNYVFTNHTWFVQSENSTEPYTDYFIWTKEQPANWVSKINMPAFTRSEKRQMFYLHQFCDNCADLNFDNPKVVEKFDMVLKAWMGAGASGVRLNNARHLLVELLEEKTRVGRGSSVDADHMRYDFWEHKHTTDLPKLKELLARWSKIVTKESEPTVFTLKEDGSLPDLILLNHNVSMLRPPSAAPVPVTEDADALASRINKTLGSWPILQLTDTNDDRELASFAMLLPAAPLLNIKQLSADENDTSLLDLVNLRSDASLSHGEHHVAAAPARNASENVLIVCARWKEGHTGYMAVYNPSRQDLHANLTAVPSVPGTITIQNVSPAVKLVTNYTKNYQPAEDVLVPAKSTVVVSYVPIKHEKREKDE
Summary
Uniprot
H9J6C6
A0A2H1WEE3
A0A2A4KA08
S4PYD3
A0A2W1BLN8
A0A194QDG9
+ More
A0A0N1PG78 A0A1Y1L2U1 A0A0C9REW8 A0A154P4V6 A0A0C9RAT0 A0A2P8Y0B1 A0A1B0GJZ3 A0A023EYE6 A0A0K8TTJ8 E2AHZ9 A0A088AB81 A0A158NL55 A0A2A3EQQ8 A0A232FDF5 A0A195BH69 K7J315 A0A195BYX0 A0A1B0AGK7 A0A1A9VYC9 E9IF95 A0A3L8D8L6 F4WFW1 A0A026WWK2 A0A1B0B6R9 A0A1I8PS72 A0A1A9Y942 A0A151XBH6 E2B6I8 A0A1A9WB84 D6WPZ5 A0A195F0G7 A0A0C9PVY1 A0A1I8PS20 A0A0C9R838 U5EQW4 A0A1I8PS64 A0A0M8ZTV6 A0A336M6J3 A0A034VX96 Q7YSY3 A0A1I8PS26 Q17I13 A0A1B0GNI1 A0A1L8DF76 A0A182GL79 E0VMF4 A0A1S4F280 A0A0K8U2N2 A0A0L7QU50 A0A151JSJ5 Q297P3 A0A023EUJ0 B4G2T8 A0A1L8DF91 A0A1B6DTJ3 A0A084WF23 A0A0R3NN61 A0A0L0CEP8 W8BJS1 B4M4Z8 A0A1B6FVL0 A0A3B0K561 T1PDH1 A0A069DVS1 A0A3B0KRG2 A0A0Q9X1K2 Q8SYR7 Q9VHX9 W8BCK4 A0A1I8M8V4 A0A0A1WNE7 A0A1Q3F9U4 B3NYT8 A0A182J069 B4NI85 B4KCN8 A0A0B4KFA6 A0A0Q9XB50 A0A1I8M8W0 B3M239 A0A182QL47 A0A1W4VZ80 B4PQY9 A0A182FJK5 A0A2M4A792 A0A2M4A764 T1E2K5
A0A0N1PG78 A0A1Y1L2U1 A0A0C9REW8 A0A154P4V6 A0A0C9RAT0 A0A2P8Y0B1 A0A1B0GJZ3 A0A023EYE6 A0A0K8TTJ8 E2AHZ9 A0A088AB81 A0A158NL55 A0A2A3EQQ8 A0A232FDF5 A0A195BH69 K7J315 A0A195BYX0 A0A1B0AGK7 A0A1A9VYC9 E9IF95 A0A3L8D8L6 F4WFW1 A0A026WWK2 A0A1B0B6R9 A0A1I8PS72 A0A1A9Y942 A0A151XBH6 E2B6I8 A0A1A9WB84 D6WPZ5 A0A195F0G7 A0A0C9PVY1 A0A1I8PS20 A0A0C9R838 U5EQW4 A0A1I8PS64 A0A0M8ZTV6 A0A336M6J3 A0A034VX96 Q7YSY3 A0A1I8PS26 Q17I13 A0A1B0GNI1 A0A1L8DF76 A0A182GL79 E0VMF4 A0A1S4F280 A0A0K8U2N2 A0A0L7QU50 A0A151JSJ5 Q297P3 A0A023EUJ0 B4G2T8 A0A1L8DF91 A0A1B6DTJ3 A0A084WF23 A0A0R3NN61 A0A0L0CEP8 W8BJS1 B4M4Z8 A0A1B6FVL0 A0A3B0K561 T1PDH1 A0A069DVS1 A0A3B0KRG2 A0A0Q9X1K2 Q8SYR7 Q9VHX9 W8BCK4 A0A1I8M8V4 A0A0A1WNE7 A0A1Q3F9U4 B3NYT8 A0A182J069 B4NI85 B4KCN8 A0A0B4KFA6 A0A0Q9XB50 A0A1I8M8W0 B3M239 A0A182QL47 A0A1W4VZ80 B4PQY9 A0A182FJK5 A0A2M4A792 A0A2M4A764 T1E2K5
Pubmed
19121390
23622113
28756777
26354079
28004739
29403074
+ More
25474469 26369729 20798317 21347285 28648823 20075255 21282665 30249741 21719571 24508170 18362917 19820115 25348373 12878228 17510324 26483478 20566863 15632085 17994087 24945155 24438588 26108605 24495485 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25830018 17550304 24330624
25474469 26369729 20798317 21347285 28648823 20075255 21282665 30249741 21719571 24508170 18362917 19820115 25348373 12878228 17510324 26483478 20566863 15632085 17994087 24945155 24438588 26108605 24495485 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25830018 17550304 24330624
EMBL
BABH01019810
ODYU01007791
SOQ50854.1
NWSH01000029
PCG80560.1
GAIX01003488
+ More
JAA89072.1 KZ150042 PZC74544.1 KQ459324 KPJ01506.1 KQ460973 KPJ10071.1 GEZM01068220 GEZM01068219 JAV67081.1 GBYB01005531 GBYB01005857 JAG75298.1 JAG75624.1 KQ434809 KZC06344.1 GBYB01013584 JAG83351.1 PYGN01001089 PSN37701.1 AJWK01022469 AJWK01022470 GBBI01004554 JAC14158.1 GDAI01000125 JAI17478.1 GL439621 EFN66907.1 ADTU01019257 KZ288194 PBC33834.1 NNAY01000371 OXU28826.1 KQ976488 KYM83516.1 KQ978501 KYM93520.1 GL762798 EFZ20756.1 QOIP01000011 RLU16631.1 GL888128 EGI66728.1 KK107078 EZA60455.1 JXJN01009222 KQ982320 KYQ57725.1 GL445975 EFN88680.1 KQ971354 EFA06898.1 KQ981866 KYN34075.1 GBYB01005638 JAG75405.1 GBYB01009042 JAG78809.1 GANO01004061 JAB55810.1 KQ435848 KOX71035.1 UFQT01000201 SSX21648.1 GAKP01012799 JAC46153.1 AY249246 AAP76306.1 CH477244 EAT46263.1 AJVK01029484 GFDF01009074 JAV05010.1 JXUM01071391 JXUM01071392 JXUM01071393 JXUM01071394 JXUM01071395 KQ562662 KXJ75373.1 DS235307 EEB14560.1 GDHF01031347 JAI20967.1 KQ414741 KOC61981.1 KQ978530 KYN30312.1 CM000070 EAL28162.2 GAPW01000518 JAC13080.1 CH479179 EDW24133.1 GFDF01009080 JAV05004.1 GEDC01008301 JAS28997.1 ATLV01023287 KE525342 KFB48817.1 KRT00512.1 JRES01000501 KNC30692.1 GAMC01007608 JAB98947.1 CH940652 EDW59709.2 GECZ01015539 JAS54230.1 OUUW01000013 SPP87832.1 KA646837 AFP61466.1 GBGD01000884 JAC88005.1 SPP87831.1 CH933806 KRG01805.1 AY071357 AAL48979.1 AE014297 BT044083 AAF54170.1 ACH92148.1 AGB95736.1 AGB95737.1 GAMC01007610 JAB98945.1 GBXI01013713 JAD00579.1 GFDL01010710 JAV24335.1 CH954181 EDV48201.1 AXCP01007513 CH964272 EDW84777.2 EDW15887.2 AGB95738.1 KRG01806.1 CH902617 EDV43363.2 AXCN02001385 CM000160 EDW96313.2 GGFK01003304 MBW36625.1 GGFK01003303 MBW36624.1 GALA01001180 JAA93672.1
JAA89072.1 KZ150042 PZC74544.1 KQ459324 KPJ01506.1 KQ460973 KPJ10071.1 GEZM01068220 GEZM01068219 JAV67081.1 GBYB01005531 GBYB01005857 JAG75298.1 JAG75624.1 KQ434809 KZC06344.1 GBYB01013584 JAG83351.1 PYGN01001089 PSN37701.1 AJWK01022469 AJWK01022470 GBBI01004554 JAC14158.1 GDAI01000125 JAI17478.1 GL439621 EFN66907.1 ADTU01019257 KZ288194 PBC33834.1 NNAY01000371 OXU28826.1 KQ976488 KYM83516.1 KQ978501 KYM93520.1 GL762798 EFZ20756.1 QOIP01000011 RLU16631.1 GL888128 EGI66728.1 KK107078 EZA60455.1 JXJN01009222 KQ982320 KYQ57725.1 GL445975 EFN88680.1 KQ971354 EFA06898.1 KQ981866 KYN34075.1 GBYB01005638 JAG75405.1 GBYB01009042 JAG78809.1 GANO01004061 JAB55810.1 KQ435848 KOX71035.1 UFQT01000201 SSX21648.1 GAKP01012799 JAC46153.1 AY249246 AAP76306.1 CH477244 EAT46263.1 AJVK01029484 GFDF01009074 JAV05010.1 JXUM01071391 JXUM01071392 JXUM01071393 JXUM01071394 JXUM01071395 KQ562662 KXJ75373.1 DS235307 EEB14560.1 GDHF01031347 JAI20967.1 KQ414741 KOC61981.1 KQ978530 KYN30312.1 CM000070 EAL28162.2 GAPW01000518 JAC13080.1 CH479179 EDW24133.1 GFDF01009080 JAV05004.1 GEDC01008301 JAS28997.1 ATLV01023287 KE525342 KFB48817.1 KRT00512.1 JRES01000501 KNC30692.1 GAMC01007608 JAB98947.1 CH940652 EDW59709.2 GECZ01015539 JAS54230.1 OUUW01000013 SPP87832.1 KA646837 AFP61466.1 GBGD01000884 JAC88005.1 SPP87831.1 CH933806 KRG01805.1 AY071357 AAL48979.1 AE014297 BT044083 AAF54170.1 ACH92148.1 AGB95736.1 AGB95737.1 GAMC01007610 JAB98945.1 GBXI01013713 JAD00579.1 GFDL01010710 JAV24335.1 CH954181 EDV48201.1 AXCP01007513 CH964272 EDW84777.2 EDW15887.2 AGB95738.1 KRG01806.1 CH902617 EDV43363.2 AXCN02001385 CM000160 EDW96313.2 GGFK01003304 MBW36625.1 GGFK01003303 MBW36624.1 GALA01001180 JAA93672.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000076502
UP000245037
+ More
UP000092461 UP000000311 UP000005203 UP000005205 UP000242457 UP000215335 UP000078540 UP000002358 UP000078542 UP000092445 UP000078200 UP000279307 UP000007755 UP000053097 UP000092460 UP000095300 UP000092443 UP000075809 UP000008237 UP000091820 UP000007266 UP000078541 UP000053105 UP000008820 UP000092462 UP000069940 UP000249989 UP000009046 UP000053825 UP000078492 UP000001819 UP000008744 UP000030765 UP000037069 UP000008792 UP000268350 UP000009192 UP000000803 UP000095301 UP000008711 UP000075880 UP000007798 UP000007801 UP000075886 UP000192221 UP000002282 UP000069272
UP000092461 UP000000311 UP000005203 UP000005205 UP000242457 UP000215335 UP000078540 UP000002358 UP000078542 UP000092445 UP000078200 UP000279307 UP000007755 UP000053097 UP000092460 UP000095300 UP000092443 UP000075809 UP000008237 UP000091820 UP000007266 UP000078541 UP000053105 UP000008820 UP000092462 UP000069940 UP000249989 UP000009046 UP000053825 UP000078492 UP000001819 UP000008744 UP000030765 UP000037069 UP000008792 UP000268350 UP000009192 UP000000803 UP000095301 UP000008711 UP000075880 UP000007798 UP000007801 UP000075886 UP000192221 UP000002282 UP000069272
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
H9J6C6
A0A2H1WEE3
A0A2A4KA08
S4PYD3
A0A2W1BLN8
A0A194QDG9
+ More
A0A0N1PG78 A0A1Y1L2U1 A0A0C9REW8 A0A154P4V6 A0A0C9RAT0 A0A2P8Y0B1 A0A1B0GJZ3 A0A023EYE6 A0A0K8TTJ8 E2AHZ9 A0A088AB81 A0A158NL55 A0A2A3EQQ8 A0A232FDF5 A0A195BH69 K7J315 A0A195BYX0 A0A1B0AGK7 A0A1A9VYC9 E9IF95 A0A3L8D8L6 F4WFW1 A0A026WWK2 A0A1B0B6R9 A0A1I8PS72 A0A1A9Y942 A0A151XBH6 E2B6I8 A0A1A9WB84 D6WPZ5 A0A195F0G7 A0A0C9PVY1 A0A1I8PS20 A0A0C9R838 U5EQW4 A0A1I8PS64 A0A0M8ZTV6 A0A336M6J3 A0A034VX96 Q7YSY3 A0A1I8PS26 Q17I13 A0A1B0GNI1 A0A1L8DF76 A0A182GL79 E0VMF4 A0A1S4F280 A0A0K8U2N2 A0A0L7QU50 A0A151JSJ5 Q297P3 A0A023EUJ0 B4G2T8 A0A1L8DF91 A0A1B6DTJ3 A0A084WF23 A0A0R3NN61 A0A0L0CEP8 W8BJS1 B4M4Z8 A0A1B6FVL0 A0A3B0K561 T1PDH1 A0A069DVS1 A0A3B0KRG2 A0A0Q9X1K2 Q8SYR7 Q9VHX9 W8BCK4 A0A1I8M8V4 A0A0A1WNE7 A0A1Q3F9U4 B3NYT8 A0A182J069 B4NI85 B4KCN8 A0A0B4KFA6 A0A0Q9XB50 A0A1I8M8W0 B3M239 A0A182QL47 A0A1W4VZ80 B4PQY9 A0A182FJK5 A0A2M4A792 A0A2M4A764 T1E2K5
A0A0N1PG78 A0A1Y1L2U1 A0A0C9REW8 A0A154P4V6 A0A0C9RAT0 A0A2P8Y0B1 A0A1B0GJZ3 A0A023EYE6 A0A0K8TTJ8 E2AHZ9 A0A088AB81 A0A158NL55 A0A2A3EQQ8 A0A232FDF5 A0A195BH69 K7J315 A0A195BYX0 A0A1B0AGK7 A0A1A9VYC9 E9IF95 A0A3L8D8L6 F4WFW1 A0A026WWK2 A0A1B0B6R9 A0A1I8PS72 A0A1A9Y942 A0A151XBH6 E2B6I8 A0A1A9WB84 D6WPZ5 A0A195F0G7 A0A0C9PVY1 A0A1I8PS20 A0A0C9R838 U5EQW4 A0A1I8PS64 A0A0M8ZTV6 A0A336M6J3 A0A034VX96 Q7YSY3 A0A1I8PS26 Q17I13 A0A1B0GNI1 A0A1L8DF76 A0A182GL79 E0VMF4 A0A1S4F280 A0A0K8U2N2 A0A0L7QU50 A0A151JSJ5 Q297P3 A0A023EUJ0 B4G2T8 A0A1L8DF91 A0A1B6DTJ3 A0A084WF23 A0A0R3NN61 A0A0L0CEP8 W8BJS1 B4M4Z8 A0A1B6FVL0 A0A3B0K561 T1PDH1 A0A069DVS1 A0A3B0KRG2 A0A0Q9X1K2 Q8SYR7 Q9VHX9 W8BCK4 A0A1I8M8V4 A0A0A1WNE7 A0A1Q3F9U4 B3NYT8 A0A182J069 B4NI85 B4KCN8 A0A0B4KFA6 A0A0Q9XB50 A0A1I8M8W0 B3M239 A0A182QL47 A0A1W4VZ80 B4PQY9 A0A182FJK5 A0A2M4A792 A0A2M4A764 T1E2K5
PDB
1M53
E-value=5.81096e-17,
Score=216
Ontologies
GO
PANTHER
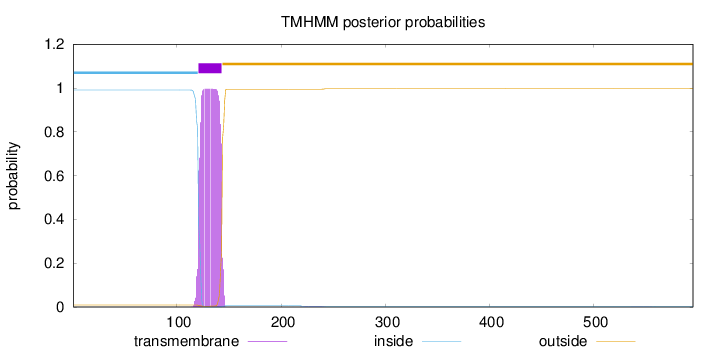
Topology
Length:
595
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.4187699999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99081
inside
1 - 120
TMhelix
121 - 143
outside
144 - 595
Population Genetic Test Statistics
Pi
236.173435
Theta
180.648523
Tajima's D
1.525803
CLR
0.295516
CSRT
0.788010599470026
Interpretation
Uncertain
