Gene
KWMTBOMO15319
Pre Gene Modal
BGIBMGA005139
Annotation
BTB/POZ_domain_containing_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.071
Sequence
CDS
ATGTCTGTAGCGCAAACCAATACTTGGATGAATGCAGAAAATATTAACAATGGTGGCGGGCTTTCCCTGTCTCCACCACACACTGTATCACAGCGAGAGACTGGTACACAGGTGTCTCAGTGCTATAGCGGGCCTCCCTCACCTTGTGGGTCATCAAGTGCGACCACGTCGCCAGCGAGTTCGCCAGCACCTCCTGGTACCGCCACCCTGGATCCCAACTGGCAAGCGACAAAGCCTACAGTTCGAGAGAGAAACGCTGCTATGTTCAATAACCAGCTAATGTCTGATATTACTTTTATTGTCGGAGCCCCAGGACACACAAAGATTATACCAGCACACAAATATGTGTTAGCAACTGCAAGTTCAGTCTTCTATGCTATGTTTTATGGAGGTTTGGCAGAATGTAAACAAGAAATTGAGGTTCCAGATGTAGAGCCATCTGCGTTCCTTGCTTTACTAAAGTATATTTACTGTGATGAAATCCAACTGGAGGCAGATACTGTATTATCAACTCTCTATGTAGCCAAAAAATATATTGTACCTCATCTGGCTAAGGCATGTGTCAATTATTTGGAAACTAGTCTCACAGCAAAAAATGCATGCCTTCTTTTAAGTCAGTCTCGACTTTTTGAGGAGCCTGAACTAATGCAGAGATGCTGGGAGGTCATAGATGCCCAGGCTGAAATGGCTTTGACGTCAGAAGGGTTTGTGGACATTGATATGTCGACATTGGAGTCAGTGTTAGCAAGGGAAACTCTGAACTGTAAAGAAATTAACTTATTTGAGTCAGCATTAGCATGGGCACATGCTGAATGTATGCGTAGAGATATAGATCCTTCACCAATTAACAAAAGAGCGATGCTTGGAAATGCCATATACTTGATCAGATTTCCCACAATGTCTTTAGAAGAATTTGCAAACAGTGCTGCTCAACTTGGAATACTAACCCCGCAAGAAACCATAGATATATTCTTGCATTTTACTGCATGCAGTAAACCAATGCTATCTTATCCCATTAAATCCAGAGCTGGATTGAAAGCTCAGATTTGCCACAGATTTCAATCATGTGCCTACAGGAGTAATCAGTGGAGGTACAGAGGACGATGTGACTCTATACAGTTCTGTGTAGATAAAAGAATATTTGTTGTTGGATTCGGACTTTATGGATCTTCAAATGGTGCAGCTGATTATAATGTTAAAATTGAATTAAAACGCCTCGGAAGAGTTCTAGCAGAGAATAATACAAAGTTCTTCTCTGATGGTTCAAGCAACACTTTTCATGTATACTTTGAAAACCCCATTCAAATTGAACCTGAATATTTCTACACTGCATCAGCAATACTAGATGGGAGTGAATTAAGCTATTTTGGACAAGAAGGCTTGAGTGAGGTGTATATGGGCACAGTGACATTCCAGTTTCACTGTTCTTCTGAGAGTACTAATGGTACTGGAGTGCAAGGAGGACAAATACCTGAACTCATTTATTATGGACCTACTATATGTGCTCCTGCTATTAAGAATTCTAGTCCAAATGAAGACTGA
Protein
MSVAQTNTWMNAENINNGGGLSLSPPHTVSQRETGTQVSQCYSGPPSPCGSSSATTSPASSPAPPGTATLDPNWQATKPTVRERNAAMFNNQLMSDITFIVGAPGHTKIIPAHKYVLATASSVFYAMFYGGLAECKQEIEVPDVEPSAFLALLKYIYCDEIQLEADTVLSTLYVAKKYIVPHLAKACVNYLETSLTAKNACLLLSQSRLFEEPELMQRCWEVIDAQAEMALTSEGFVDIDMSTLESVLARETLNCKEINLFESALAWAHAECMRRDIDPSPINKRAMLGNAIYLIRFPTMSLEEFANSAAQLGILTPQETIDIFLHFTACSKPMLSYPIKSRAGLKAQICHRFQSCAYRSNQWRYRGRCDSIQFCVDKRIFVVGFGLYGSSNGAADYNVKIELKRLGRVLAENNTKFFSDGSSNTFHVYFENPIQIEPEYFYTASAILDGSELSYFGQEGLSEVYMGTVTFQFHCSSESTNGTGVQGGQIPELIYYGPTICAPAIKNSSPNED
Summary
Uniprot
H9J6J7
Q2F626
A0A2A4J7F2
A0A2H1V725
G9BC28
A0A2A4J656
+ More
A0A212F910 A0A0L7KQL9 A0A2W1BER6 A0A194PHA7 A0A194RKY1 A0A3S2LPH5 A0A0T6B906 A0A2J7RIK5 A0A2P8XGT9 D6WSI4 A0A067RKV5 A0A1Y1LE33 A0A1W4WH21 A0A1W4WI32 A0A2J7RIK4 A0A1B6EA09 U4UDP7 A0A1Y1LGC1 A0A1B6IEI6 A0A1B6KSK7 A0A1B6DYY6 A0A1W4WIB4 A0A1W4WI40 A0A1W4W6N9 A0A1W4W6N6 A0A1W4WIB8 A0A1W4W6N1 A0A1W4WH26 A0A1W4WIC3 E0VUM9 A0A1B6K0P8 A0A1B6LYV3 A0A0V0G5N6 N6TNL2 A0A069DVP9 A0A0P4VUF4 A0A232F5X4 E2AAF1 A0A224XN86 A0A336LUJ3 A0A026WWR0 A0A2A3ECG9 A0A088ACY4 T1I4P2 A0A195CCP3 A0A151WNN8 U5ETU9 A0A195DVN3 F4WAW3 A0A151I3M1 A0A158NSM7 A0A1L8DB12 A0A310SPA0 A0A182Y5E3 A0A182Q6D5 Q7QB19 A0A182XG50 A0A182ID96 A0A182V935 Q17IA8 A0A182RV64 A0A182PD77 A0A182ND05 A0A182W1A7 A0A1S4F1N4 A0A2M4CZL0 A0A1Q3G413 A0A1Q3G492 A0A1Q3FUB9 A0A1B0CP17 A0A1Q3FUK8 A0A2M4BEZ4 A0A2M4BEU9 A0A2M4CPP1 A0A2M4CQ40 A0A2M3Z1X0 A0A2M4A2V9 A0A2M4A2R1 A0A1J1I776 A0A2M3Z1I0 W5JNY3 A0A2M3Z1I8 A0A2M4A2S6 A0A182FPU3 T1IIE9 A0A1Q3G434 A0A1Q3G450 A0A0A9XFQ0 A0A1Q3G495 A0A1Q3FUQ0 A0A1Q3FUL5 A0A1Q3FUD9 A0A1Q3FU98 A0A084VBG4
A0A212F910 A0A0L7KQL9 A0A2W1BER6 A0A194PHA7 A0A194RKY1 A0A3S2LPH5 A0A0T6B906 A0A2J7RIK5 A0A2P8XGT9 D6WSI4 A0A067RKV5 A0A1Y1LE33 A0A1W4WH21 A0A1W4WI32 A0A2J7RIK4 A0A1B6EA09 U4UDP7 A0A1Y1LGC1 A0A1B6IEI6 A0A1B6KSK7 A0A1B6DYY6 A0A1W4WIB4 A0A1W4WI40 A0A1W4W6N9 A0A1W4W6N6 A0A1W4WIB8 A0A1W4W6N1 A0A1W4WH26 A0A1W4WIC3 E0VUM9 A0A1B6K0P8 A0A1B6LYV3 A0A0V0G5N6 N6TNL2 A0A069DVP9 A0A0P4VUF4 A0A232F5X4 E2AAF1 A0A224XN86 A0A336LUJ3 A0A026WWR0 A0A2A3ECG9 A0A088ACY4 T1I4P2 A0A195CCP3 A0A151WNN8 U5ETU9 A0A195DVN3 F4WAW3 A0A151I3M1 A0A158NSM7 A0A1L8DB12 A0A310SPA0 A0A182Y5E3 A0A182Q6D5 Q7QB19 A0A182XG50 A0A182ID96 A0A182V935 Q17IA8 A0A182RV64 A0A182PD77 A0A182ND05 A0A182W1A7 A0A1S4F1N4 A0A2M4CZL0 A0A1Q3G413 A0A1Q3G492 A0A1Q3FUB9 A0A1B0CP17 A0A1Q3FUK8 A0A2M4BEZ4 A0A2M4BEU9 A0A2M4CPP1 A0A2M4CQ40 A0A2M3Z1X0 A0A2M4A2V9 A0A2M4A2R1 A0A1J1I776 A0A2M3Z1I0 W5JNY3 A0A2M3Z1I8 A0A2M4A2S6 A0A182FPU3 T1IIE9 A0A1Q3G434 A0A1Q3G450 A0A0A9XFQ0 A0A1Q3G495 A0A1Q3FUQ0 A0A1Q3FUL5 A0A1Q3FUD9 A0A1Q3FU98 A0A084VBG4
Pubmed
EMBL
BABH01019830
DQ311246
ABD36191.1
NWSH01002741
PCG67598.1
ODYU01000766
+ More
SOQ36054.1 HQ331539 ADO32579.1 PCG67597.1 AGBW02009658 OWR50211.1 JTDY01006908 KOB65598.1 KZ150095 PZC73612.1 KQ459604 KPI92433.1 KQ460045 KPJ18074.1 RSAL01000466 RVE41624.1 LJIG01009111 KRT83733.1 NEVH01003500 PNF40664.1 PYGN01002190 PSN31220.1 KQ971352 EFA06364.2 KK852408 KDR24501.1 GEZM01058097 JAV71909.1 PNF40665.1 GEDC01002532 JAS34766.1 KB632233 ERL90423.1 GEZM01058098 JAV71908.1 GECU01026839 GECU01022372 JAS80867.1 JAS85334.1 GEBQ01025572 GEBQ01003557 JAT14405.1 JAT36420.1 GEDC01006414 JAS30884.1 DS235787 EEB17085.1 GECU01035912 GECU01002667 JAS71794.1 JAT05040.1 GEBQ01017941 GEBQ01011104 JAT22036.1 JAT28873.1 GECL01002763 JAP03361.1 APGK01057875 KB741282 ENN70845.1 GBGD01001107 JAC87782.1 GDKW01000278 JAI56317.1 NNAY01000907 OXU25982.1 GL438113 EFN69586.1 GFTR01006813 JAW09613.1 UFQT01000205 SSX21702.1 KK107078 QOIP01000011 EZA60497.1 RLU16844.1 KZ288285 PBC29473.1 ACPB03014599 KQ978023 KYM97991.1 KQ982906 KYQ49484.1 GANO01002619 JAB57252.1 KQ980322 KYN16619.1 GL888054 EGI68653.1 KQ976482 KYM83704.1 ADTU01025038 ADTU01025039 ADTU01025040 GFDF01010580 JAV03504.1 KQ760551 OAD59823.1 AXCN02001566 AAAB01008880 EAA08648.4 APCN01005054 CH477241 EAT46385.1 GGFL01006537 MBW70715.1 GFDL01000511 JAV34534.1 GFDL01000425 JAV34620.1 GFDL01003890 JAV31155.1 AJWK01021315 GFDL01003879 JAV31166.1 GGFJ01002446 MBW51587.1 GGFJ01002445 MBW51586.1 GGFL01003122 MBW67300.1 GGFL01003123 MBW67301.1 GGFM01001752 MBW22503.1 GGFK01001741 MBW35062.1 GGFK01001746 MBW35067.1 CVRI01000041 CRK95436.1 GGFM01001622 MBW22373.1 ADMH02000944 ETN64605.1 GGFM01001624 MBW22375.1 GGFK01001748 MBW35069.1 AFFK01014262 GFDL01000482 JAV34563.1 GFDL01000467 JAV34578.1 GBHO01027679 GDHC01021042 JAG15925.1 JAP97586.1 GFDL01000422 JAV34623.1 GFDL01003872 JAV31173.1 GFDL01003869 JAV31176.1 GFDL01003870 JAV31175.1 GFDL01003871 JAV31174.1 ATLV01006809 KE524386 KFB35308.1
SOQ36054.1 HQ331539 ADO32579.1 PCG67597.1 AGBW02009658 OWR50211.1 JTDY01006908 KOB65598.1 KZ150095 PZC73612.1 KQ459604 KPI92433.1 KQ460045 KPJ18074.1 RSAL01000466 RVE41624.1 LJIG01009111 KRT83733.1 NEVH01003500 PNF40664.1 PYGN01002190 PSN31220.1 KQ971352 EFA06364.2 KK852408 KDR24501.1 GEZM01058097 JAV71909.1 PNF40665.1 GEDC01002532 JAS34766.1 KB632233 ERL90423.1 GEZM01058098 JAV71908.1 GECU01026839 GECU01022372 JAS80867.1 JAS85334.1 GEBQ01025572 GEBQ01003557 JAT14405.1 JAT36420.1 GEDC01006414 JAS30884.1 DS235787 EEB17085.1 GECU01035912 GECU01002667 JAS71794.1 JAT05040.1 GEBQ01017941 GEBQ01011104 JAT22036.1 JAT28873.1 GECL01002763 JAP03361.1 APGK01057875 KB741282 ENN70845.1 GBGD01001107 JAC87782.1 GDKW01000278 JAI56317.1 NNAY01000907 OXU25982.1 GL438113 EFN69586.1 GFTR01006813 JAW09613.1 UFQT01000205 SSX21702.1 KK107078 QOIP01000011 EZA60497.1 RLU16844.1 KZ288285 PBC29473.1 ACPB03014599 KQ978023 KYM97991.1 KQ982906 KYQ49484.1 GANO01002619 JAB57252.1 KQ980322 KYN16619.1 GL888054 EGI68653.1 KQ976482 KYM83704.1 ADTU01025038 ADTU01025039 ADTU01025040 GFDF01010580 JAV03504.1 KQ760551 OAD59823.1 AXCN02001566 AAAB01008880 EAA08648.4 APCN01005054 CH477241 EAT46385.1 GGFL01006537 MBW70715.1 GFDL01000511 JAV34534.1 GFDL01000425 JAV34620.1 GFDL01003890 JAV31155.1 AJWK01021315 GFDL01003879 JAV31166.1 GGFJ01002446 MBW51587.1 GGFJ01002445 MBW51586.1 GGFL01003122 MBW67300.1 GGFL01003123 MBW67301.1 GGFM01001752 MBW22503.1 GGFK01001741 MBW35062.1 GGFK01001746 MBW35067.1 CVRI01000041 CRK95436.1 GGFM01001622 MBW22373.1 ADMH02000944 ETN64605.1 GGFM01001624 MBW22375.1 GGFK01001748 MBW35069.1 AFFK01014262 GFDL01000482 JAV34563.1 GFDL01000467 JAV34578.1 GBHO01027679 GDHC01021042 JAG15925.1 JAP97586.1 GFDL01000422 JAV34623.1 GFDL01003872 JAV31173.1 GFDL01003869 JAV31176.1 GFDL01003870 JAV31175.1 GFDL01003871 JAV31174.1 ATLV01006809 KE524386 KFB35308.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053268
UP000053240
+ More
UP000283053 UP000235965 UP000245037 UP000007266 UP000027135 UP000192223 UP000030742 UP000009046 UP000019118 UP000215335 UP000000311 UP000053097 UP000279307 UP000242457 UP000005203 UP000015103 UP000078542 UP000075809 UP000078492 UP000007755 UP000078540 UP000005205 UP000076408 UP000075886 UP000007062 UP000076407 UP000075840 UP000075903 UP000008820 UP000075900 UP000075885 UP000075884 UP000075920 UP000092461 UP000183832 UP000000673 UP000069272 UP000030765
UP000283053 UP000235965 UP000245037 UP000007266 UP000027135 UP000192223 UP000030742 UP000009046 UP000019118 UP000215335 UP000000311 UP000053097 UP000279307 UP000242457 UP000005203 UP000015103 UP000078542 UP000075809 UP000078492 UP000007755 UP000078540 UP000005205 UP000076408 UP000075886 UP000007062 UP000076407 UP000075840 UP000075903 UP000008820 UP000075900 UP000075885 UP000075884 UP000075920 UP000092461 UP000183832 UP000000673 UP000069272 UP000030765
Interpro
SUPFAM
SSF54695
SSF54695
Gene 3D
ProteinModelPortal
H9J6J7
Q2F626
A0A2A4J7F2
A0A2H1V725
G9BC28
A0A2A4J656
+ More
A0A212F910 A0A0L7KQL9 A0A2W1BER6 A0A194PHA7 A0A194RKY1 A0A3S2LPH5 A0A0T6B906 A0A2J7RIK5 A0A2P8XGT9 D6WSI4 A0A067RKV5 A0A1Y1LE33 A0A1W4WH21 A0A1W4WI32 A0A2J7RIK4 A0A1B6EA09 U4UDP7 A0A1Y1LGC1 A0A1B6IEI6 A0A1B6KSK7 A0A1B6DYY6 A0A1W4WIB4 A0A1W4WI40 A0A1W4W6N9 A0A1W4W6N6 A0A1W4WIB8 A0A1W4W6N1 A0A1W4WH26 A0A1W4WIC3 E0VUM9 A0A1B6K0P8 A0A1B6LYV3 A0A0V0G5N6 N6TNL2 A0A069DVP9 A0A0P4VUF4 A0A232F5X4 E2AAF1 A0A224XN86 A0A336LUJ3 A0A026WWR0 A0A2A3ECG9 A0A088ACY4 T1I4P2 A0A195CCP3 A0A151WNN8 U5ETU9 A0A195DVN3 F4WAW3 A0A151I3M1 A0A158NSM7 A0A1L8DB12 A0A310SPA0 A0A182Y5E3 A0A182Q6D5 Q7QB19 A0A182XG50 A0A182ID96 A0A182V935 Q17IA8 A0A182RV64 A0A182PD77 A0A182ND05 A0A182W1A7 A0A1S4F1N4 A0A2M4CZL0 A0A1Q3G413 A0A1Q3G492 A0A1Q3FUB9 A0A1B0CP17 A0A1Q3FUK8 A0A2M4BEZ4 A0A2M4BEU9 A0A2M4CPP1 A0A2M4CQ40 A0A2M3Z1X0 A0A2M4A2V9 A0A2M4A2R1 A0A1J1I776 A0A2M3Z1I0 W5JNY3 A0A2M3Z1I8 A0A2M4A2S6 A0A182FPU3 T1IIE9 A0A1Q3G434 A0A1Q3G450 A0A0A9XFQ0 A0A1Q3G495 A0A1Q3FUQ0 A0A1Q3FUL5 A0A1Q3FUD9 A0A1Q3FU98 A0A084VBG4
A0A212F910 A0A0L7KQL9 A0A2W1BER6 A0A194PHA7 A0A194RKY1 A0A3S2LPH5 A0A0T6B906 A0A2J7RIK5 A0A2P8XGT9 D6WSI4 A0A067RKV5 A0A1Y1LE33 A0A1W4WH21 A0A1W4WI32 A0A2J7RIK4 A0A1B6EA09 U4UDP7 A0A1Y1LGC1 A0A1B6IEI6 A0A1B6KSK7 A0A1B6DYY6 A0A1W4WIB4 A0A1W4WI40 A0A1W4W6N9 A0A1W4W6N6 A0A1W4WIB8 A0A1W4W6N1 A0A1W4WH26 A0A1W4WIC3 E0VUM9 A0A1B6K0P8 A0A1B6LYV3 A0A0V0G5N6 N6TNL2 A0A069DVP9 A0A0P4VUF4 A0A232F5X4 E2AAF1 A0A224XN86 A0A336LUJ3 A0A026WWR0 A0A2A3ECG9 A0A088ACY4 T1I4P2 A0A195CCP3 A0A151WNN8 U5ETU9 A0A195DVN3 F4WAW3 A0A151I3M1 A0A158NSM7 A0A1L8DB12 A0A310SPA0 A0A182Y5E3 A0A182Q6D5 Q7QB19 A0A182XG50 A0A182ID96 A0A182V935 Q17IA8 A0A182RV64 A0A182PD77 A0A182ND05 A0A182W1A7 A0A1S4F1N4 A0A2M4CZL0 A0A1Q3G413 A0A1Q3G492 A0A1Q3FUB9 A0A1B0CP17 A0A1Q3FUK8 A0A2M4BEZ4 A0A2M4BEU9 A0A2M4CPP1 A0A2M4CQ40 A0A2M3Z1X0 A0A2M4A2V9 A0A2M4A2R1 A0A1J1I776 A0A2M3Z1I0 W5JNY3 A0A2M3Z1I8 A0A2M4A2S6 A0A182FPU3 T1IIE9 A0A1Q3G434 A0A1Q3G450 A0A0A9XFQ0 A0A1Q3G495 A0A1Q3FUQ0 A0A1Q3FUL5 A0A1Q3FUD9 A0A1Q3FU98 A0A084VBG4
PDB
2VKP
E-value=5.65935e-44,
Score=448
Ontologies
KEGG
GO
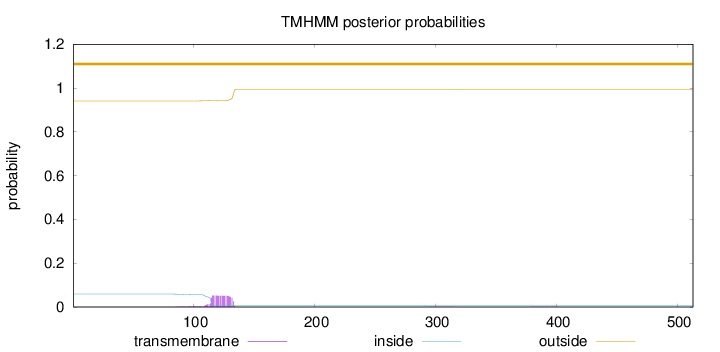
Topology
Length:
513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.05325
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.05912
outside
1 - 513
Population Genetic Test Statistics
Pi
179.821067
Theta
16.53514
Tajima's D
0.446778
CLR
0
CSRT
0.507174641267937
Interpretation
Uncertain
