Gene
KWMTBOMO15317
Pre Gene Modal
BGIBMGA005140
Annotation
PREDICTED:_transcription_factor_kayak?_isoforms_A/B/F_isoform_X3_[Papilio_xuthus]
Transcription factor
Location in the cell
Nuclear Reliability : 4.609
Sequence
CDS
ATGCAGAATATCGATCCTTTGGAGATCGCGAACATCCTCGCTACCGAGCTCTGGTGTCAGCAGTTGGCAAACCTCGAAGGACTCCAGTCAGGAGTCCCGACCCGCACGACGGCGACCATCACCCCCACTCAGTTGCGGAACTTCGAGCAGACGTACATCGAGTTGACCAACTGCCGAAGCGAACCAACCACTCACGTCGGCTTCGTACCACCTTCAGTCACGCACGCCAACAATTACGGTATTCTTAATTCGGTGGGATACTGTGATTCGGGCCCGACGACCGCGCTGCACGTGTCGCCGGGCCCGCTCTCCGCTAGCGGCGACAGCAGCAGCAGCCCTGGTCTCCCGGCGCCGAAACGACGCAACATGGGCGGTCGTAGACCGAACAAAGCACCGCAGGAACTATCGCCCGAAGAGGAAGAACGAAGAAAGATACGCCGCGAAAGGAACAAGATGGCGGCCGCGCGGTGTCGCAAACGCAGACTCGATCACACTAACGAACTGCAAGACGAGACCGATAGATTGGAGCAAAAGAGACAGTCGCTCCAAGAGGAAATAAGAAAGTTAAACGCGGACCGGGAGCACCTTCAAACGATATTGCAGAACCATATGATAACGTGCCGGCTCAGCGACCGCTCTTCGAGTCCGCCGGACGTGAAGCCCCTCCAGGAAGTTTACACGTATCCGGACCCGGAGGACGGAGTTCGAGTCAAAACCGAAGTCATAGACCCCACAGTCGACCCAGTAATGCTGGGGTTGGATAACATTTTCACAACACCGTCGACCGAAAAAAGGATAATGCTGTCTTCGGCCAACCCTGGCGTGGTGACGAGTGGGGATACGGTCACGTTGGATACCCCGCCCGCTATTTCTCGACCCAATAGGCCTAATTCACTACAAGTACCGCTCAGTCTCACACCTGCACAGATACACAACAACAAAGTACTCGGGAACAGCAAAATAGCCGGCATCGAGATCAGCACGCCTAGCAACGGCATGTTTAACTACGAAAGCCTCATGGACGGGGGCACGGGGTTGACCCCAGTGCTTCCCCACCCGCTAGGACACGCGCACGCGTACGTGGTCCCCCCGCGCCCCGCCCCGCCCGGCGCCGCCGACCCCCCCGAGCTGGTGTCGCTGTGA
Protein
MQNIDPLEIANILATELWCQQLANLEGLQSGVPTRTTATITPTQLRNFEQTYIELTNCRSEPTTHVGFVPPSVTHANNYGILNSVGYCDSGPTTALHVSPGPLSASGDSSSSPGLPAPKRRNMGGRRPNKAPQELSPEEEERRKIRRERNKMAAARCRKRRLDHTNELQDETDRLEQKRQSLQEEIRKLNADREHLQTILQNHMITCRLSDRSSSPPDVKPLQEVYTYPDPEDGVRVKTEVIDPTVDPVMLGLDNIFTTPSTEKRIMLSSANPGVVTSGDTVTLDTPPAISRPNRPNSLQVPLSLTPAQIHNNKVLGNSKIAGIEISTPSNGMFNYESLMDGGTGLTPVLPHPLGHAHAYVVPPRPAPPGAADPPELVSL
Summary
Uniprot
H9J6J8
A0A2H1VLZ5
A0A2A4J689
A0A212F930
A0A194RJS4
A0A194PI76
+ More
A0A1E1W590 A0A2A4J877 A0A2A4J861 A0A1E1WC41 A0A2W1BIV1 A0A0L7L5X4 A0A2A4J7U5 A0A1W4X605 E1ZVL5 F4W793 A0A195BJW8 A0A1Y1MEQ9 A0A2J7R6I7 E2BK04 A0A026VYK7 A0A1W7HGV6 A0A2P8YXE8 E0VJ27 V5GRA9 A0A2J7R6I0 D6WZA9 A0A1Y1MAU3 A0A0L7QM80 A0A0J7P0U3 A0A158NJ42 A0A151WQA7 A0A0C9S042 A0A195EY11 V5GV79 A0A1B6D3U9 A0A067QPW4 A0A3L8DCT0 A0A154P5Z3 A0A195DIP5 A0A1B6D2H0 N6ULZ4 U5EYJ9 A0A1W7R9H3 A0A0A9X176 A0A2I9LP45 A0A0A9ZHE2 A0A0A9Z8S4 A0A0A9XP53 A0A146KT21 A0A146M4L0 A0A0P4VRY3 A0A1A9XMV9 A0A1A9UQ64 A0A1B0BZ81 A0A1B0G287 A0A1B0A8W4 A0A1A9W6I6 A0A2R5L8J3 A0A0L0BMT9
A0A1E1W590 A0A2A4J877 A0A2A4J861 A0A1E1WC41 A0A2W1BIV1 A0A0L7L5X4 A0A2A4J7U5 A0A1W4X605 E1ZVL5 F4W793 A0A195BJW8 A0A1Y1MEQ9 A0A2J7R6I7 E2BK04 A0A026VYK7 A0A1W7HGV6 A0A2P8YXE8 E0VJ27 V5GRA9 A0A2J7R6I0 D6WZA9 A0A1Y1MAU3 A0A0L7QM80 A0A0J7P0U3 A0A158NJ42 A0A151WQA7 A0A0C9S042 A0A195EY11 V5GV79 A0A1B6D3U9 A0A067QPW4 A0A3L8DCT0 A0A154P5Z3 A0A195DIP5 A0A1B6D2H0 N6ULZ4 U5EYJ9 A0A1W7R9H3 A0A0A9X176 A0A2I9LP45 A0A0A9ZHE2 A0A0A9Z8S4 A0A0A9XP53 A0A146KT21 A0A146M4L0 A0A0P4VRY3 A0A1A9XMV9 A0A1A9UQ64 A0A1B0BZ81 A0A1B0G287 A0A1B0A8W4 A0A1A9W6I6 A0A2R5L8J3 A0A0L0BMT9
Pubmed
EMBL
BABH01019830
ODYU01003116
SOQ41462.1
NWSH01002741
PCG67595.1
AGBW02009658
+ More
OWR50209.1 KQ460045 KPJ18078.1 KQ459604 KPI92429.1 GDQN01008906 JAT82148.1 PCG67593.1 PCG67592.1 GDQN01006508 JAT84546.1 KZ150095 PZC73614.1 JTDY01002716 KOB70862.1 PCG67594.1 GL434548 EFN74784.1 GL887813 EGI69977.1 KQ976453 KYM85468.1 GEZM01036309 JAV82815.1 NEVH01006756 PNF36440.1 GL448708 EFN84013.1 KK107746 EZA47944.1 LC215243 BAX36487.1 PYGN01000302 PSN48912.1 DS235219 EEB13383.1 GALX01004329 JAB64137.1 PNF36438.1 KQ971372 EFA10699.1 GEZM01036310 JAV82814.1 KQ414894 KOC59753.1 LBMM01000467 KMQ98265.1 ADTU01017434 ADTU01017435 ADTU01017436 ADTU01017437 ADTU01017438 KQ982845 KYQ49988.1 GBYB01013786 JAG83553.1 KQ981920 KYN33036.1 GALX01004328 JAB64138.1 GEDC01016950 JAS20348.1 KK853153 KDR10584.1 QOIP01000010 RLU17728.1 KQ434809 KZC06618.1 KQ980804 KYN12768.1 GEDC01017452 JAS19846.1 APGK01018358 KB740073 ENN81681.1 GANO01000310 JAB59561.1 GFAH01000614 JAV47775.1 GBHO01038964 GBHO01030182 GBHO01030181 GBHO01030178 GBHO01018873 GBHO01018871 GDHC01002070 JAG04640.1 JAG13422.1 JAG13423.1 JAG13426.1 JAG24731.1 JAG24733.1 JAQ16559.1 GFWZ01000167 MBW20157.1 GBHO01039021 GBHO01035092 GBHO01034769 GBHO01022207 GBHO01001978 GBHO01001974 JAG04583.1 JAG08512.1 JAG08835.1 JAG21397.1 JAG41626.1 JAG41630.1 GBHO01039045 GBHO01038961 GBHO01030183 GBHO01012090 GBHO01001979 GBHO01001977 JAG04559.1 JAG04643.1 JAG13421.1 JAG31514.1 JAG41625.1 JAG41627.1 GBHO01030184 GBHO01030180 GBHO01030179 GBHO01022183 GBHO01001980 GBHO01001976 JAG13420.1 JAG13424.1 JAG13425.1 JAG21421.1 JAG41624.1 JAG41628.1 GDHC01019530 JAP99098.1 GDHC01012599 GDHC01005129 JAQ06030.1 JAQ13500.1 GDRN01102556 JAI58223.1 JXJN01023015 CCAG010011799 CCAG010011800 CCAG010011801 CCAG010011802 CCAG010011803 CCAG010011804 GGLE01001673 MBY05799.1 JRES01001623 KNC21405.1
OWR50209.1 KQ460045 KPJ18078.1 KQ459604 KPI92429.1 GDQN01008906 JAT82148.1 PCG67593.1 PCG67592.1 GDQN01006508 JAT84546.1 KZ150095 PZC73614.1 JTDY01002716 KOB70862.1 PCG67594.1 GL434548 EFN74784.1 GL887813 EGI69977.1 KQ976453 KYM85468.1 GEZM01036309 JAV82815.1 NEVH01006756 PNF36440.1 GL448708 EFN84013.1 KK107746 EZA47944.1 LC215243 BAX36487.1 PYGN01000302 PSN48912.1 DS235219 EEB13383.1 GALX01004329 JAB64137.1 PNF36438.1 KQ971372 EFA10699.1 GEZM01036310 JAV82814.1 KQ414894 KOC59753.1 LBMM01000467 KMQ98265.1 ADTU01017434 ADTU01017435 ADTU01017436 ADTU01017437 ADTU01017438 KQ982845 KYQ49988.1 GBYB01013786 JAG83553.1 KQ981920 KYN33036.1 GALX01004328 JAB64138.1 GEDC01016950 JAS20348.1 KK853153 KDR10584.1 QOIP01000010 RLU17728.1 KQ434809 KZC06618.1 KQ980804 KYN12768.1 GEDC01017452 JAS19846.1 APGK01018358 KB740073 ENN81681.1 GANO01000310 JAB59561.1 GFAH01000614 JAV47775.1 GBHO01038964 GBHO01030182 GBHO01030181 GBHO01030178 GBHO01018873 GBHO01018871 GDHC01002070 JAG04640.1 JAG13422.1 JAG13423.1 JAG13426.1 JAG24731.1 JAG24733.1 JAQ16559.1 GFWZ01000167 MBW20157.1 GBHO01039021 GBHO01035092 GBHO01034769 GBHO01022207 GBHO01001978 GBHO01001974 JAG04583.1 JAG08512.1 JAG08835.1 JAG21397.1 JAG41626.1 JAG41630.1 GBHO01039045 GBHO01038961 GBHO01030183 GBHO01012090 GBHO01001979 GBHO01001977 JAG04559.1 JAG04643.1 JAG13421.1 JAG31514.1 JAG41625.1 JAG41627.1 GBHO01030184 GBHO01030180 GBHO01030179 GBHO01022183 GBHO01001980 GBHO01001976 JAG13420.1 JAG13424.1 JAG13425.1 JAG21421.1 JAG41624.1 JAG41628.1 GDHC01019530 JAP99098.1 GDHC01012599 GDHC01005129 JAQ06030.1 JAQ13500.1 GDRN01102556 JAI58223.1 JXJN01023015 CCAG010011799 CCAG010011800 CCAG010011801 CCAG010011802 CCAG010011803 CCAG010011804 GGLE01001673 MBY05799.1 JRES01001623 KNC21405.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000192223 UP000000311 UP000007755 UP000078540 UP000235965 UP000008237 UP000053097 UP000245037 UP000009046 UP000007266 UP000053825 UP000036403 UP000005205 UP000075809 UP000078541 UP000027135 UP000279307 UP000076502 UP000078492 UP000019118 UP000092443 UP000078200 UP000092460 UP000092444 UP000092445 UP000091820 UP000037069
UP000192223 UP000000311 UP000007755 UP000078540 UP000235965 UP000008237 UP000053097 UP000245037 UP000009046 UP000007266 UP000053825 UP000036403 UP000005205 UP000075809 UP000078541 UP000027135 UP000279307 UP000076502 UP000078492 UP000019118 UP000092443 UP000078200 UP000092460 UP000092444 UP000092445 UP000091820 UP000037069
PRIDE
ProteinModelPortal
H9J6J8
A0A2H1VLZ5
A0A2A4J689
A0A212F930
A0A194RJS4
A0A194PI76
+ More
A0A1E1W590 A0A2A4J877 A0A2A4J861 A0A1E1WC41 A0A2W1BIV1 A0A0L7L5X4 A0A2A4J7U5 A0A1W4X605 E1ZVL5 F4W793 A0A195BJW8 A0A1Y1MEQ9 A0A2J7R6I7 E2BK04 A0A026VYK7 A0A1W7HGV6 A0A2P8YXE8 E0VJ27 V5GRA9 A0A2J7R6I0 D6WZA9 A0A1Y1MAU3 A0A0L7QM80 A0A0J7P0U3 A0A158NJ42 A0A151WQA7 A0A0C9S042 A0A195EY11 V5GV79 A0A1B6D3U9 A0A067QPW4 A0A3L8DCT0 A0A154P5Z3 A0A195DIP5 A0A1B6D2H0 N6ULZ4 U5EYJ9 A0A1W7R9H3 A0A0A9X176 A0A2I9LP45 A0A0A9ZHE2 A0A0A9Z8S4 A0A0A9XP53 A0A146KT21 A0A146M4L0 A0A0P4VRY3 A0A1A9XMV9 A0A1A9UQ64 A0A1B0BZ81 A0A1B0G287 A0A1B0A8W4 A0A1A9W6I6 A0A2R5L8J3 A0A0L0BMT9
A0A1E1W590 A0A2A4J877 A0A2A4J861 A0A1E1WC41 A0A2W1BIV1 A0A0L7L5X4 A0A2A4J7U5 A0A1W4X605 E1ZVL5 F4W793 A0A195BJW8 A0A1Y1MEQ9 A0A2J7R6I7 E2BK04 A0A026VYK7 A0A1W7HGV6 A0A2P8YXE8 E0VJ27 V5GRA9 A0A2J7R6I0 D6WZA9 A0A1Y1MAU3 A0A0L7QM80 A0A0J7P0U3 A0A158NJ42 A0A151WQA7 A0A0C9S042 A0A195EY11 V5GV79 A0A1B6D3U9 A0A067QPW4 A0A3L8DCT0 A0A154P5Z3 A0A195DIP5 A0A1B6D2H0 N6ULZ4 U5EYJ9 A0A1W7R9H3 A0A0A9X176 A0A2I9LP45 A0A0A9ZHE2 A0A0A9Z8S4 A0A0A9XP53 A0A146KT21 A0A146M4L0 A0A0P4VRY3 A0A1A9XMV9 A0A1A9UQ64 A0A1B0BZ81 A0A1B0G287 A0A1B0A8W4 A0A1A9W6I6 A0A2R5L8J3 A0A0L0BMT9
PDB
2WT7
E-value=6.89784e-08,
Score=136
Ontologies
PATHWAY
GO
PANTHER
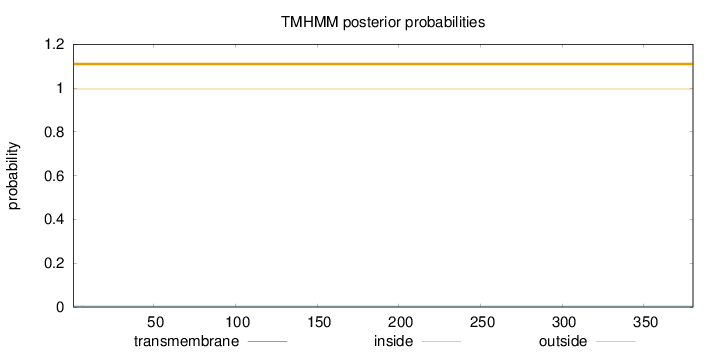
Topology
Length:
380
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000870000000000001
Exp number, first 60 AAs:
0.00056
Total prob of N-in:
0.00492
outside
1 - 380
Population Genetic Test Statistics
Pi
173.177435
Theta
171.3033
Tajima's D
0.17076
CLR
0.747623
CSRT
0.415179241037948
Interpretation
Uncertain
