Gene
KWMTBOMO15316
Annotation
PREDICTED:_sodium_channel_protein_Nach-like_[Bombyx_mori]
Full name
Sodium channel protein Nach
Alternative Name
Pickpocket protein 4
Location in the cell
PlasmaMembrane Reliability : 2.053
Sequence
CDS
ATGCGATCAGCCGCTTGCAACTGTCCAAGAGATTGTGAGTCACGCCAGTACAAAGTTGACATTAGCACTGGGAATTTGAACGCTCTGCCGTACATTCCTAACAATCCCTTTGCAGATGTAACCTTCAAGCGTAGTACATCAATCATGCGTTTCATTTTTCCAAACTCCGTATACGTGAAACAGAAGCAAGAAACCGTCGTGCCTTTGATCAGCCTCGTCTCTAATCTCGGAGGCGTTTTCGGTCTATGTTTGGGTTGTAGCTGCATCAGCGTGCTCGAAATATTATTCTTTTCATATCTGTATATTAAGAGAAAAATTCGTAAGCACTTTATTAACCCACGTAAGTGA
Protein
MRSAACNCPRDCESRQYKVDISTGNLNALPYIPNNPFADVTFKRSTSIMRFIFPNSVYVKQKQETVVPLISLVSNLGGVFGLCLGCSCISVLEILFFSYLYIKRKIRKHFINPRK
Summary
Description
Part of a complex that plays a role in tracheal liquid clearance. Probable role in sodium transport (By similarity).
Part of a complex that plays a role in tracheal liquid clearance. Probable role in sodium transport.
Part of a complex that plays a role in tracheal liquid clearance. Probable role in sodium transport.
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Keywords
Glycoprotein
Ion channel
Ion transport
Membrane
Sodium
Sodium channel
Sodium transport
Transmembrane
Transmembrane helix
Transport
Alternative splicing
Complete proteome
Reference proteome
Feature
chain Sodium channel protein Nach
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A194RKY6
A0A194PHA1
A0A232FJV4
K7J1Q6
A0A067QUS0
A0A182RBD3
+ More
Q7PRJ3 Q28WP2 B4H760 A0A182UQ91 A0A182K6C4 A0A182W3S6 A0A182LY59 A0A182YS00 A0A182NDK8 A0A1I8P290 A0A0L7RDT9 A0A182GCJ0 A0A1B0AWZ8 A0A182PVF2 A0A182QFT1 A0A1J1J2M5 A0A084WQY4 E2A4M1 A0A336M002 A0A195B3V5 A0A3L8DSJ1 W5JM38 A0A3B0IYE0 A0A182GCJ1 A0A336MTW7 A0A139WFX5 A0A0K8VFG4 A0A0P8XQJ1 A0A182GXU6 A0A0P9AG67 B3MDJ9 B4MIU3 A0A1W4UXC8 A0A1I8PG23 A0A1W4XB49 Q17MX5 A0A1I8PAZ4 B4LP99 Q9NJP2 O61370 B4QIE2 A0A336KF41 A0A1I8Q569 A0A182GRG8 B4P5X5 A0A182FM32 A0A1I8N7J9 A0A158Q786 A0A238BZ76 A0A0J7KTG4 A0A183HZK6 A0A182NA42 T1H366 A0A1I8M339 A0A0N4W7B7 B4HT74 A0A1S4EX71 A0A1S4FSE9 A0A182FLG5 Q16RC1 K7IP12 A0A1I8PEH0 A0A1I8Q523 A0A232FIR5 A0A1S4EW59 E2BM03 A0A182KL79 A0A1I8P9G1 A0A1I8P9B5 B4JWN2 B3NPF9 B0WDC2 W2T696 A0A1A9Z9F9 A0A384TM68 O61365 O61365-3 A0A384SX96 A0A0L0BVZ9 A0A194PW83 A0A1W5C9C1 O61365-2 B4NYM0 A0A182E6X6 J9F3I1
Q7PRJ3 Q28WP2 B4H760 A0A182UQ91 A0A182K6C4 A0A182W3S6 A0A182LY59 A0A182YS00 A0A182NDK8 A0A1I8P290 A0A0L7RDT9 A0A182GCJ0 A0A1B0AWZ8 A0A182PVF2 A0A182QFT1 A0A1J1J2M5 A0A084WQY4 E2A4M1 A0A336M002 A0A195B3V5 A0A3L8DSJ1 W5JM38 A0A3B0IYE0 A0A182GCJ1 A0A336MTW7 A0A139WFX5 A0A0K8VFG4 A0A0P8XQJ1 A0A182GXU6 A0A0P9AG67 B3MDJ9 B4MIU3 A0A1W4UXC8 A0A1I8PG23 A0A1W4XB49 Q17MX5 A0A1I8PAZ4 B4LP99 Q9NJP2 O61370 B4QIE2 A0A336KF41 A0A1I8Q569 A0A182GRG8 B4P5X5 A0A182FM32 A0A1I8N7J9 A0A158Q786 A0A238BZ76 A0A0J7KTG4 A0A183HZK6 A0A182NA42 T1H366 A0A1I8M339 A0A0N4W7B7 B4HT74 A0A1S4EX71 A0A1S4FSE9 A0A182FLG5 Q16RC1 K7IP12 A0A1I8PEH0 A0A1I8Q523 A0A232FIR5 A0A1S4EW59 E2BM03 A0A182KL79 A0A1I8P9G1 A0A1I8P9B5 B4JWN2 B3NPF9 B0WDC2 W2T696 A0A1A9Z9F9 A0A384TM68 O61365 O61365-3 A0A384SX96 A0A0L0BVZ9 A0A194PW83 A0A1W5C9C1 O61365-2 B4NYM0 A0A182E6X6 J9F3I1
Pubmed
EMBL
KQ460045
KPJ18079.1
KQ459604
KPI92428.1
NNAY01000116
OXU30820.1
+ More
KK853243 KDR09460.1 AAAB01008849 EAA07138.5 CM000071 EAL26625.3 CH479216 EDW33593.1 AXCM01000168 KQ414612 KOC69152.1 JXUM01054263 KQ561813 KXJ77446.1 JXJN01004965 AXCN02001756 CVRI01000066 CRL06218.1 ATLV01025839 KE525401 KFB52628.1 GL436710 EFN71623.1 UFQT01000343 SSX23390.1 KQ976625 KYM78952.1 QOIP01000005 RLU22829.1 ADMH02000731 ETN65216.1 OUUW01000001 SPP72777.1 KXJ77447.1 UFQT01001968 SSX32233.1 KQ971351 KYB26842.1 GDHF01014716 JAI37598.1 CH902619 KPU76840.1 JXUM01096280 JXUM01096281 JXUM01096282 KQ564253 KXJ72525.1 KPU76839.1 EDV37462.1 CH963719 EDW72032.1 CH477203 EAT48027.1 CH940648 EDW60208.1 AF136603 AAF61426.1 AF024691 CM000362 CM002911 EDX07389.1 KMY94317.1 UFQS01000390 UFQT01000390 SSX03496.1 SSX23861.1 JXUM01082415 KQ563311 KXJ74083.1 CM000158 EDW91891.1 KZ269987 OZC10394.1 LBMM01003440 KMQ93534.1 UZAJ01039976 VDP12491.1 CAQQ02171020 UZAF01016422 VDO27681.1 CH480816 EDW48175.1 CH477717 EAT36963.1 NNAY01000169 OXU30358.1 GL449100 EFN83252.1 CH916375 EDV98370.1 CH954179 EDV55726.1 DS231896 EDS44435.1 KI660223 ETN76502.1 AE013599 API64972.1 AF022713 AY226538 AAO47364.1 API64973.1 JRES01001352 KNC23384.1 KQ459591 KPI97019.1 AAAB01008823 EAU77465.2 CM000157 EDW88684.1 UYRW01000759 VDK70442.1 ADBV01003284 EJW81894.1
KK853243 KDR09460.1 AAAB01008849 EAA07138.5 CM000071 EAL26625.3 CH479216 EDW33593.1 AXCM01000168 KQ414612 KOC69152.1 JXUM01054263 KQ561813 KXJ77446.1 JXJN01004965 AXCN02001756 CVRI01000066 CRL06218.1 ATLV01025839 KE525401 KFB52628.1 GL436710 EFN71623.1 UFQT01000343 SSX23390.1 KQ976625 KYM78952.1 QOIP01000005 RLU22829.1 ADMH02000731 ETN65216.1 OUUW01000001 SPP72777.1 KXJ77447.1 UFQT01001968 SSX32233.1 KQ971351 KYB26842.1 GDHF01014716 JAI37598.1 CH902619 KPU76840.1 JXUM01096280 JXUM01096281 JXUM01096282 KQ564253 KXJ72525.1 KPU76839.1 EDV37462.1 CH963719 EDW72032.1 CH477203 EAT48027.1 CH940648 EDW60208.1 AF136603 AAF61426.1 AF024691 CM000362 CM002911 EDX07389.1 KMY94317.1 UFQS01000390 UFQT01000390 SSX03496.1 SSX23861.1 JXUM01082415 KQ563311 KXJ74083.1 CM000158 EDW91891.1 KZ269987 OZC10394.1 LBMM01003440 KMQ93534.1 UZAJ01039976 VDP12491.1 CAQQ02171020 UZAF01016422 VDO27681.1 CH480816 EDW48175.1 CH477717 EAT36963.1 NNAY01000169 OXU30358.1 GL449100 EFN83252.1 CH916375 EDV98370.1 CH954179 EDV55726.1 DS231896 EDS44435.1 KI660223 ETN76502.1 AE013599 API64972.1 AF022713 AY226538 AAO47364.1 API64973.1 JRES01001352 KNC23384.1 KQ459591 KPI97019.1 AAAB01008823 EAU77465.2 CM000157 EDW88684.1 UYRW01000759 VDK70442.1 ADBV01003284 EJW81894.1
Proteomes
UP000053240
UP000053268
UP000215335
UP000002358
UP000027135
UP000075900
+ More
UP000007062 UP000001819 UP000008744 UP000075903 UP000075881 UP000075920 UP000075883 UP000076408 UP000075884 UP000095300 UP000053825 UP000069940 UP000249989 UP000092460 UP000075885 UP000075886 UP000183832 UP000030765 UP000000311 UP000078540 UP000279307 UP000000673 UP000268350 UP000007266 UP000007801 UP000007798 UP000192221 UP000192223 UP000008820 UP000008792 UP000000304 UP000002282 UP000069272 UP000095301 UP000050640 UP000036403 UP000050787 UP000015102 UP000038042 UP000268014 UP000001292 UP000008237 UP000075882 UP000001070 UP000008711 UP000002320 UP000092445 UP000000803 UP000037069 UP000077448 UP000271087 UP000004810
UP000007062 UP000001819 UP000008744 UP000075903 UP000075881 UP000075920 UP000075883 UP000076408 UP000075884 UP000095300 UP000053825 UP000069940 UP000249989 UP000092460 UP000075885 UP000075886 UP000183832 UP000030765 UP000000311 UP000078540 UP000279307 UP000000673 UP000268350 UP000007266 UP000007801 UP000007798 UP000192221 UP000192223 UP000008820 UP000008792 UP000000304 UP000002282 UP000069272 UP000095301 UP000050640 UP000036403 UP000050787 UP000015102 UP000038042 UP000268014 UP000001292 UP000008237 UP000075882 UP000001070 UP000008711 UP000002320 UP000092445 UP000000803 UP000037069 UP000077448 UP000271087 UP000004810
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A194RKY6
A0A194PHA1
A0A232FJV4
K7J1Q6
A0A067QUS0
A0A182RBD3
+ More
Q7PRJ3 Q28WP2 B4H760 A0A182UQ91 A0A182K6C4 A0A182W3S6 A0A182LY59 A0A182YS00 A0A182NDK8 A0A1I8P290 A0A0L7RDT9 A0A182GCJ0 A0A1B0AWZ8 A0A182PVF2 A0A182QFT1 A0A1J1J2M5 A0A084WQY4 E2A4M1 A0A336M002 A0A195B3V5 A0A3L8DSJ1 W5JM38 A0A3B0IYE0 A0A182GCJ1 A0A336MTW7 A0A139WFX5 A0A0K8VFG4 A0A0P8XQJ1 A0A182GXU6 A0A0P9AG67 B3MDJ9 B4MIU3 A0A1W4UXC8 A0A1I8PG23 A0A1W4XB49 Q17MX5 A0A1I8PAZ4 B4LP99 Q9NJP2 O61370 B4QIE2 A0A336KF41 A0A1I8Q569 A0A182GRG8 B4P5X5 A0A182FM32 A0A1I8N7J9 A0A158Q786 A0A238BZ76 A0A0J7KTG4 A0A183HZK6 A0A182NA42 T1H366 A0A1I8M339 A0A0N4W7B7 B4HT74 A0A1S4EX71 A0A1S4FSE9 A0A182FLG5 Q16RC1 K7IP12 A0A1I8PEH0 A0A1I8Q523 A0A232FIR5 A0A1S4EW59 E2BM03 A0A182KL79 A0A1I8P9G1 A0A1I8P9B5 B4JWN2 B3NPF9 B0WDC2 W2T696 A0A1A9Z9F9 A0A384TM68 O61365 O61365-3 A0A384SX96 A0A0L0BVZ9 A0A194PW83 A0A1W5C9C1 O61365-2 B4NYM0 A0A182E6X6 J9F3I1
Q7PRJ3 Q28WP2 B4H760 A0A182UQ91 A0A182K6C4 A0A182W3S6 A0A182LY59 A0A182YS00 A0A182NDK8 A0A1I8P290 A0A0L7RDT9 A0A182GCJ0 A0A1B0AWZ8 A0A182PVF2 A0A182QFT1 A0A1J1J2M5 A0A084WQY4 E2A4M1 A0A336M002 A0A195B3V5 A0A3L8DSJ1 W5JM38 A0A3B0IYE0 A0A182GCJ1 A0A336MTW7 A0A139WFX5 A0A0K8VFG4 A0A0P8XQJ1 A0A182GXU6 A0A0P9AG67 B3MDJ9 B4MIU3 A0A1W4UXC8 A0A1I8PG23 A0A1W4XB49 Q17MX5 A0A1I8PAZ4 B4LP99 Q9NJP2 O61370 B4QIE2 A0A336KF41 A0A1I8Q569 A0A182GRG8 B4P5X5 A0A182FM32 A0A1I8N7J9 A0A158Q786 A0A238BZ76 A0A0J7KTG4 A0A183HZK6 A0A182NA42 T1H366 A0A1I8M339 A0A0N4W7B7 B4HT74 A0A1S4EX71 A0A1S4FSE9 A0A182FLG5 Q16RC1 K7IP12 A0A1I8PEH0 A0A1I8Q523 A0A232FIR5 A0A1S4EW59 E2BM03 A0A182KL79 A0A1I8P9G1 A0A1I8P9B5 B4JWN2 B3NPF9 B0WDC2 W2T696 A0A1A9Z9F9 A0A384TM68 O61365 O61365-3 A0A384SX96 A0A0L0BVZ9 A0A194PW83 A0A1W5C9C1 O61365-2 B4NYM0 A0A182E6X6 J9F3I1
PDB
4NYK
E-value=0.058138,
Score=78
Ontologies
GO
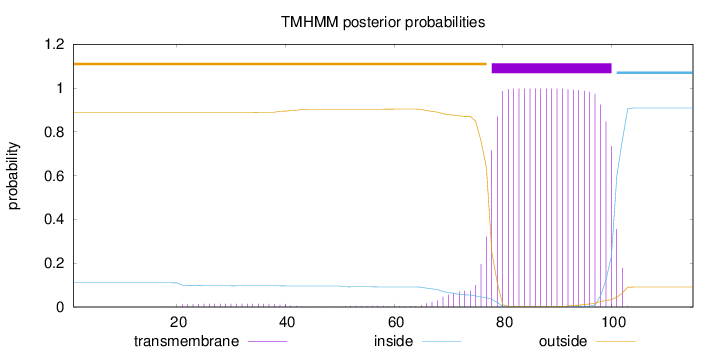
Topology
Subcellular location
Membrane
Length:
115
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.93546
Exp number, first 60 AAs:
0.33704
Total prob of N-in:
0.11100
outside
1 - 77
TMhelix
78 - 100
inside
101 - 115
Population Genetic Test Statistics
Pi
209.815617
Theta
20.968953
Tajima's D
0.297405
CLR
56.671003
CSRT
0.459077046147693
Interpretation
Uncertain
