Gene
KWMTBOMO15314 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005058
Annotation
Na?K-ATPase_alpha-subunit_[Bombyx_mori]
Full name
Sodium/potassium-transporting ATPase subunit alpha
Alternative Name
Sodium pump subunit alpha
Location in the cell
PlasmaMembrane Reliability : 3.589
Sequence
CDS
ATGGGCGAGACGCGCAGAAAACCTCCCGCGAAAAAACGGAAACCAGGAGATTTAGAAGATCTCAAACAGGAGCTGGATATTGATTACCATAAAGTCACTCCTGAGGAACTCTACCAAAGATTTCAAACGCACCCGGAAAATGGCCTTAGTCACGCAAAAGCGAAAGAGAACTTAGAAAGAGACGGCCCCAATGCTCTTACACCACCTAAACAAACACCCGAATGGGTGAAGTTTTGTAAAAATCTATTCGGCGGCTTTGCATTGTTACTCTGGATTGGTGCTATTCTATGCTTTATCGCCTACGGTATTCAGGCTAGTACCGTAGAGGAGCCCGCTGATGACAATCTGTACCTGGGCATCGTGCTCGCGGCCGTCGTGATCGTTACTGGTATCTTCTCGTACTACCAAGAGAGCAAGTCGTCTAAAATCATGGAATCTTTCAAGAACATGGTCCCCCAATTCGCCACCGTCATCCGTGAAGGAGAGAAACTTACGCTCCGAGCCGAGGACCTGGTGCTTGGTGATATTGTCGAGGTTAAATTCGGTGACCGGATTCCCGCCGACATCCGTATTATCGAAGCGCGAGGCTTTAAAGTCGACAACTCTAGCTTGACCGGCGAATCTGAACCCCAATCCCGAGGAGCCGAATTCACAAATGAGAACCCACTCGAGACCAAGAACTTGGCCTTCTTCTCGACCAACGCCGTTGAGGGTACCGCTAAAGGAATAGTCATCTGCTGTGGAGACAATACTGTGATGGGCCGCATTGCTGGTCTCGCTTCCGGTCTGGACACTGGTGAAACTCCGATCGCTAAGGAGATCCATCACTTTATTCATCTTATCACCGGCGTGGCTGTCTTCCTCGGAGTGACTTTCTTCGTTATTGCTTTCATTCTCGGATACCACTGGCTCGACGCTGTCATCTTCCTCATCGGTATCATCGTAGCTAACGTGCCTGAAGGGCTCTTGGCCACCGTGACTGTGTGCTTGACTCTCACTGCCAAACGAATGGCTTCCAAGAACTGCCTGGTCAAGAATCTCGAAGCCGTCGAAACTCTGGGTTCCACTTCCACGATTTGCTCCGACAAAACCGGAACCCTCACTCAGAACAGAATGACTGTTGCTCACATGTGGTTCGACAACCAGATCATTGAAGCTGATACAACTGAAGATCAATCCGGCGTACAGTACGACCGCACTAGCCCTGGCTTCAAGGCTCTTGCCAAAATTGCTTCGCTCTGCAACCGAGCTGAGTTCAAGGGTGGTCAGGATGGTGTTCCGATTCTGAAGAAGGAAGTCGCCGGTGACGCCTCTGAAGCCGCTTTACTGAAATGCATGGAGCTGGCTCTCGGTGATGTCTTGTCGATCAGGAAGCGCAACAAGAAAGTCTGCGAAATTCCCTTCAACTCTACTAACAAGTACCAGGTATCGATCCACGAGAGCGACGACCCCAGCGACCCCCGCCACCTGCTGGTGATGAAGGGAGCACCCGAAAGAATCCTGGAACGTTGTAGCACTATCTTCATTGGCGGAAAGGAAAAGGTATTAGACGAGGAAATGAAGGAGGCGTTCAACAACGCTTACCTTGAACTCGGTGGTCTTGGCGAGCGTGTGCTCGGCTTCTGCGACCTTCAGCTGCCCTCCGACAAGTACCCCATCGGCTACAAGTTCAACACTGACGACCCTAACTTCCCCTTGGACAACTTGCGCTTTGTTGGTCTGATGAGCATGATTGATCCTCCCCGCGCTGCTGTACCAGATGCCGTGGCCAAGTGCCGCTCCGCCGGTATCAAGGTTATCATGGTAACCGGAGACCACCCCATCACTGCCAAGGCTATTGCCAAATCTGTAGGTATAATTTCTGAAGGTAACGAGACCGTAGAGGACATTGCTGCGCGCTTGAATATCCCCGTTTCCGAGGTGAATCCCCGCGAAGCCAAAGCCGCTGTCGTCCACGGAACCGAGCTCCGCGAACTGAACTCCGACCAGCTCGATGAGATCCTCAAGTTCCACACCGAAATCGTGTTCGCCCGCACCTCCCCGCAACAGAAGTTGATCATCGTTGAGGGTTGCCAGCGTCTCGGCGCCATCGTGGCTGTGACCGGCGACGGTGTCAACGACTCGCCCGCCCTCAAGAAGGCCGACATCGGTGTCGCTATGGGTATCGCCGGCTCCGACGTTTCCAAGCAGGCCGCTGACATGATTCTACTCGACGACAACTTCGCGTCCATCGTGACCGGTGTCGAAGAAGGCCGACTGATCTTTGACAACTTGAAGAAGTCAATCGCCTACACCCTAACCTCGAACATTCCAGAAATCTCCCCCTTCCTTGCCTTCATCCTTTGCGACATTCCTCTGCCCCTGGGTACAGTAACCATCCTCTGCATCGATCTGGGAACGGACATGGTGCCTGCTATATCATTGGCGTACGAGGCGCCCGAGTCTGATATCATGAAGCGGCAGCCGCGGGACCCTTACCGCGACAACCTCGTCAACAGAAGGTTGATCTCTATGGCTTACGGACAAATCGGAATGATCCAAGCCGCGGCTGGCTTCTTCGTCTACTTCGTGATCATGGCTGAAAACGGATTCCTCCCGATGAAGCTTTTCGGTATCAGGAAGCAGTGGGACTCGAAGGCCATCAACGATTTGACTGACTCCTATGGACAGGAATGGACCTACCGTGACCGCAAGGCGCTCGAGTTTACTTGCCACACCGCATTCTTCGTATCTATTGTCGTTGTGCAATGGGCCGACTTGATTATCTGCAAGACCCGCCGTAACTCGATCATTCACCAGGGCATGCGCAACTGGGCCCTCAACTTTGGTCTCATATTTGAAACCGCGCTGGCGGCGTTCCTCTCGTACACTCCCGGTATGGACAAGGGCTTGAGGATGTACCCACTCAAGTTCGTGTGGTGGCTGCCCGCCATTCCGTTCATGCTGTCCATCTTCATCTACGACGAGATCAGGCGCTTCTACCTGCGCCGCAACCCCGGCGGCTGGCTCGAACAAGAGACTTATTATTAA
Protein
MGETRRKPPAKKRKPGDLEDLKQELDIDYHKVTPEELYQRFQTHPENGLSHAKAKENLERDGPNALTPPKQTPEWVKFCKNLFGGFALLLWIGAILCFIAYGIQASTVEEPADDNLYLGIVLAAVVIVTGIFSYYQESKSSKIMESFKNMVPQFATVIREGEKLTLRAEDLVLGDIVEVKFGDRIPADIRIIEARGFKVDNSSLTGESEPQSRGAEFTNENPLETKNLAFFSTNAVEGTAKGIVICCGDNTVMGRIAGLASGLDTGETPIAKEIHHFIHLITGVAVFLGVTFFVIAFILGYHWLDAVIFLIGIIVANVPEGLLATVTVCLTLTAKRMASKNCLVKNLEAVETLGSTSTICSDKTGTLTQNRMTVAHMWFDNQIIEADTTEDQSGVQYDRTSPGFKALAKIASLCNRAEFKGGQDGVPILKKEVAGDASEAALLKCMELALGDVLSIRKRNKKVCEIPFNSTNKYQVSIHESDDPSDPRHLLVMKGAPERILERCSTIFIGGKEKVLDEEMKEAFNNAYLELGGLGERVLGFCDLQLPSDKYPIGYKFNTDDPNFPLDNLRFVGLMSMIDPPRAAVPDAVAKCRSAGIKVIMVTGDHPITAKAIAKSVGIISEGNETVEDIAARLNIPVSEVNPREAKAAVVHGTELRELNSDQLDEILKFHTEIVFARTSPQQKLIIVEGCQRLGAIVAVTGDGVNDSPALKKADIGVAMGIAGSDVSKQAADMILLDDNFASIVTGVEEGRLIFDNLKKSIAYTLTSNIPEISPFLAFILCDIPLPLGTVTILCIDLGTDMVPAISLAYEAPESDIMKRQPRDPYRDNLVNRRLISMAYGQIGMIQAAAGFFVYFVIMAENGFLPMKLFGIRKQWDSKAINDLTDSYGQEWTYRDRKALEFTCHTAFFVSIVVVQWADLIICKTRRNSIIHQGMRNWALNFGLIFETALAAFLSYTPGMDKGLRMYPLKFVWWLPAIPFMLSIFIYDEIRRFYLRRNPGGWLEQETYY
Summary
Description
This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients.
Catalytic Activity
ATP + H2O + K(+)(out) + Na(+)(in) = ADP + H(+) + K(+)(in) + Na(+)(out) + phosphate
Subunit
The sodium/potassium-transporting ATPase is composed of a catalytic alpha subunit, an auxiliary non-catalytic beta subunit and an additional regulatory subunit.
Miscellaneous
Ouabain-sensitive electrogenic ion pump.
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IIC subfamily.
Keywords
Alternative splicing
ATP-binding
Cell membrane
Complete proteome
Ion transport
Membrane
Nucleotide-binding
Phosphoprotein
Potassium
Potassium transport
Reference proteome
RNA editing
Sodium
Sodium transport
Translocase
Transmembrane
Transmembrane helix
Transport
Feature
chain Sodium/potassium-transporting ATPase subunit alpha
splice variant In isoform 2, isoform 3, isoform 6 and isoform 7.
splice variant In isoform 2, isoform 3, isoform 6 and isoform 7.
Uniprot
A0A0K2S4Y7
K0G7C8
K0GEX0
K0GM01
A0A2A4JVC3
K0GEX4
+ More
K0GG88 A0A2W1BK20 A0A2A4JV07 A0A2A4JUC5 A0A2H1VI27 K0GLY7 K0GG86 K0GEX7 K0G7C4 K0G7B9 A0A158NIT2 Q7PGN1 A0A1W4X4P7 K0GLY2 A0A182QT13 A0A182XW64 A0A1L8DEW0 A0A182PSN6 A0A3L8D359 V9I6B8 N6T2P0 A0A2A3E444 U4UIT1 E9IZS4 A0A158NIT1 E2AIZ4 F5HKJ1 F5HKJ2 A0A1Q3G4G9 A0A1Q3FUU5 A0A182LGE6 A0A0K8TRD8 A0A182IU13 F5HKJ4 A0A151IUE3 A0A1W4WV71 A0A0Q9X358 A0A0C9R0I4 A0A1L8DF50 A0A0Q9WBH9 Q27766 A0A1W4X5W5 A0A2M3ZZE0 A0A1W4WUY0 A0A1W4WV42 I5ANS6 A0A1Y0AWP8 A0A1W4X5V6 A0A2M4CSX9 A0A2M4CUW4 A0A182RQ21 A0A1W4X5S4 A0A2M4A020 A0A2M4A3F2 A0A1L8DEU4 A0A2M4CUE1 A0A1W4X5Q8 A0A1W4WV39 K0GG97 A0A2M4CIP8 A0A182N737 A0A0M4F6Q7 A0A0Q9X2S3 A0A026W561 A0A1I8MBT4 A0A0R1E150 A0A087ZTA6 A0A1I8PAR6 A0A0A1X4H1 A0A182VYR9 A0A1J1J616 A0A0N8P1E9 A0A2M3ZZK0 Q16N74 P13607-7 T1E1Y4 A0A0R1E1E2 A0A1J1J6V9 A0A0A1WY87 K0GLX7 A0A1Q3FV87 V9I6A9 K0GEZ1 A0A0Q9XCU5 A0A0Q9WBZ0 A0A1Q3FV30 A0A1Q3G4J5 K0GEX8 K0GEY8 A0A0K8UY58 A0A034W1V5 A0A1Q3FV07 A0A1B6MM45 A0A0Q9W0Q7
K0GG88 A0A2W1BK20 A0A2A4JV07 A0A2A4JUC5 A0A2H1VI27 K0GLY7 K0GG86 K0GEX7 K0G7C4 K0G7B9 A0A158NIT2 Q7PGN1 A0A1W4X4P7 K0GLY2 A0A182QT13 A0A182XW64 A0A1L8DEW0 A0A182PSN6 A0A3L8D359 V9I6B8 N6T2P0 A0A2A3E444 U4UIT1 E9IZS4 A0A158NIT1 E2AIZ4 F5HKJ1 F5HKJ2 A0A1Q3G4G9 A0A1Q3FUU5 A0A182LGE6 A0A0K8TRD8 A0A182IU13 F5HKJ4 A0A151IUE3 A0A1W4WV71 A0A0Q9X358 A0A0C9R0I4 A0A1L8DF50 A0A0Q9WBH9 Q27766 A0A1W4X5W5 A0A2M3ZZE0 A0A1W4WUY0 A0A1W4WV42 I5ANS6 A0A1Y0AWP8 A0A1W4X5V6 A0A2M4CSX9 A0A2M4CUW4 A0A182RQ21 A0A1W4X5S4 A0A2M4A020 A0A2M4A3F2 A0A1L8DEU4 A0A2M4CUE1 A0A1W4X5Q8 A0A1W4WV39 K0GG97 A0A2M4CIP8 A0A182N737 A0A0M4F6Q7 A0A0Q9X2S3 A0A026W561 A0A1I8MBT4 A0A0R1E150 A0A087ZTA6 A0A1I8PAR6 A0A0A1X4H1 A0A182VYR9 A0A1J1J616 A0A0N8P1E9 A0A2M3ZZK0 Q16N74 P13607-7 T1E1Y4 A0A0R1E1E2 A0A1J1J6V9 A0A0A1WY87 K0GLX7 A0A1Q3FV87 V9I6A9 K0GEZ1 A0A0Q9XCU5 A0A0Q9WBZ0 A0A1Q3FV30 A0A1Q3G4J5 K0GEX8 K0GEY8 A0A0K8UY58 A0A034W1V5 A0A1Q3FV07 A0A1B6MM45 A0A0Q9W0Q7
EC Number
7.2.2.13
Pubmed
EMBL
LC029030
BAS22117.1
JQ771512
AFU25681.1
JQ771498
AFU25667.1
+ More
JQ771525 AFU25694.1 NWSH01000562 PCG75638.1 JQ771506 AFU25675.1 JQ771509 AFU25678.1 KZ150095 PZC73617.1 PCG75639.1 PCG75637.1 ODYU01002679 SOQ40480.1 JQ771510 AFU25679.1 JQ771504 AFU25673.1 JQ771508 AFU25677.1 JQ771507 AFU25676.1 JQ771502 AFU25671.1 ADTU01017127 ADTU01017128 AAAB01008859 EAA44868.4 JQ771505 AFU25674.1 AXCN02000326 GFDF01009103 JAV04981.1 QOIP01000014 RLU14937.1 JR035472 JR035478 AEY56435.1 AEY56441.1 APGK01054932 APGK01054933 KB741253 ENN71823.1 KZ288428 PBC25851.1 KB632340 ERL92967.1 GL767232 EFZ13954.1 GL439921 EFN66583.1 EGK96744.1 EGK96743.1 GFDL01000352 JAV34693.1 GFDL01003664 JAV31381.1 GDAI01000902 JAI16701.1 EGK96746.1 KQ980960 KYN11056.1 CH933806 KRG02333.1 GBYB01000331 JAG70098.1 GFDF01009104 JAV04980.1 CH940652 KRF78692.1 S66043 AAB28239.1 GGFK01000592 MBW33913.1 CM000070 EIM52611.1 KY921817 ART29406.1 GGFL01004278 MBW68456.1 GGFL01004964 MBW69142.1 GGFK01000778 MBW34099.1 GGFK01001996 MBW35317.1 GFDF01009105 JAV04979.1 GGFL01004280 MBW68458.1 JQ771524 AFU25693.1 GGFL01001054 MBW65232.1 CP012526 ALC47775.1 CH964232 KRF99409.1 KK107405 EZA51212.1 CM000160 KRK02982.1 GBXI01008719 JAD05573.1 CVRI01000070 CRL07212.1 CH902617 KPU79732.1 GGFK01000581 MBW33902.1 CH477835 EAT35802.1 X14476 AF044974 AE014297 AY069184 AY174097 AY174098 U55767 X17471 GALA01001249 JAA93603.1 KRK02985.1 CRL07214.1 GBXI01010944 JAD03348.1 JQ771500 AFU25669.1 GFDL01003659 JAV31386.1 JR035470 JR035477 AEY56433.1 AEY56440.1 JQ771526 AFU25695.1 KRG02334.1 KRF78687.1 GFDL01003657 JAV31388.1 GFDL01000313 JAV34732.1 JQ771511 AFU25680.1 JQ771523 AFU25692.1 GDHF01020773 JAI31541.1 GAKP01010640 GAKP01010638 JAC48312.1 GFDL01003660 JAV31385.1 GEBQ01003038 JAT36939.1 KRF78691.1
JQ771525 AFU25694.1 NWSH01000562 PCG75638.1 JQ771506 AFU25675.1 JQ771509 AFU25678.1 KZ150095 PZC73617.1 PCG75639.1 PCG75637.1 ODYU01002679 SOQ40480.1 JQ771510 AFU25679.1 JQ771504 AFU25673.1 JQ771508 AFU25677.1 JQ771507 AFU25676.1 JQ771502 AFU25671.1 ADTU01017127 ADTU01017128 AAAB01008859 EAA44868.4 JQ771505 AFU25674.1 AXCN02000326 GFDF01009103 JAV04981.1 QOIP01000014 RLU14937.1 JR035472 JR035478 AEY56435.1 AEY56441.1 APGK01054932 APGK01054933 KB741253 ENN71823.1 KZ288428 PBC25851.1 KB632340 ERL92967.1 GL767232 EFZ13954.1 GL439921 EFN66583.1 EGK96744.1 EGK96743.1 GFDL01000352 JAV34693.1 GFDL01003664 JAV31381.1 GDAI01000902 JAI16701.1 EGK96746.1 KQ980960 KYN11056.1 CH933806 KRG02333.1 GBYB01000331 JAG70098.1 GFDF01009104 JAV04980.1 CH940652 KRF78692.1 S66043 AAB28239.1 GGFK01000592 MBW33913.1 CM000070 EIM52611.1 KY921817 ART29406.1 GGFL01004278 MBW68456.1 GGFL01004964 MBW69142.1 GGFK01000778 MBW34099.1 GGFK01001996 MBW35317.1 GFDF01009105 JAV04979.1 GGFL01004280 MBW68458.1 JQ771524 AFU25693.1 GGFL01001054 MBW65232.1 CP012526 ALC47775.1 CH964232 KRF99409.1 KK107405 EZA51212.1 CM000160 KRK02982.1 GBXI01008719 JAD05573.1 CVRI01000070 CRL07212.1 CH902617 KPU79732.1 GGFK01000581 MBW33902.1 CH477835 EAT35802.1 X14476 AF044974 AE014297 AY069184 AY174097 AY174098 U55767 X17471 GALA01001249 JAA93603.1 KRK02985.1 CRL07214.1 GBXI01010944 JAD03348.1 JQ771500 AFU25669.1 GFDL01003659 JAV31386.1 JR035470 JR035477 AEY56433.1 AEY56440.1 JQ771526 AFU25695.1 KRG02334.1 KRF78687.1 GFDL01003657 JAV31388.1 GFDL01000313 JAV34732.1 JQ771511 AFU25680.1 JQ771523 AFU25692.1 GDHF01020773 JAI31541.1 GAKP01010640 GAKP01010638 JAC48312.1 GFDL01003660 JAV31385.1 GEBQ01003038 JAT36939.1 KRF78691.1
Proteomes
UP000218220
UP000005205
UP000007062
UP000192223
UP000075886
UP000076408
+ More
UP000075885 UP000279307 UP000019118 UP000242457 UP000030742 UP000000311 UP000075882 UP000075880 UP000078492 UP000009192 UP000008792 UP000001819 UP000075900 UP000075884 UP000092553 UP000007798 UP000053097 UP000095301 UP000002282 UP000005203 UP000095300 UP000075920 UP000183832 UP000007801 UP000008820 UP000000803
UP000075885 UP000279307 UP000019118 UP000242457 UP000030742 UP000000311 UP000075882 UP000075880 UP000078492 UP000009192 UP000008792 UP000001819 UP000075900 UP000075884 UP000092553 UP000007798 UP000053097 UP000095301 UP000002282 UP000005203 UP000095300 UP000075920 UP000183832 UP000007801 UP000008820 UP000000803
PRIDE
Interpro
IPR018303
ATPase_P-typ_P_site
+ More
IPR004014 ATPase_P-typ_cation-transptr_N
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006068 ATPase_P-typ_cation-transptr_C
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR005775 P-type_ATPase_IIC
IPR001757 P_typ_ATPase
IPR023214 HAD_sf
IPR036412 HAD-like_sf
IPR004014 ATPase_P-typ_cation-transptr_N
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006068 ATPase_P-typ_cation-transptr_C
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR005775 P-type_ATPase_IIC
IPR001757 P_typ_ATPase
IPR023214 HAD_sf
IPR036412 HAD-like_sf
Gene 3D
ProteinModelPortal
A0A0K2S4Y7
K0G7C8
K0GEX0
K0GM01
A0A2A4JVC3
K0GEX4
+ More
K0GG88 A0A2W1BK20 A0A2A4JV07 A0A2A4JUC5 A0A2H1VI27 K0GLY7 K0GG86 K0GEX7 K0G7C4 K0G7B9 A0A158NIT2 Q7PGN1 A0A1W4X4P7 K0GLY2 A0A182QT13 A0A182XW64 A0A1L8DEW0 A0A182PSN6 A0A3L8D359 V9I6B8 N6T2P0 A0A2A3E444 U4UIT1 E9IZS4 A0A158NIT1 E2AIZ4 F5HKJ1 F5HKJ2 A0A1Q3G4G9 A0A1Q3FUU5 A0A182LGE6 A0A0K8TRD8 A0A182IU13 F5HKJ4 A0A151IUE3 A0A1W4WV71 A0A0Q9X358 A0A0C9R0I4 A0A1L8DF50 A0A0Q9WBH9 Q27766 A0A1W4X5W5 A0A2M3ZZE0 A0A1W4WUY0 A0A1W4WV42 I5ANS6 A0A1Y0AWP8 A0A1W4X5V6 A0A2M4CSX9 A0A2M4CUW4 A0A182RQ21 A0A1W4X5S4 A0A2M4A020 A0A2M4A3F2 A0A1L8DEU4 A0A2M4CUE1 A0A1W4X5Q8 A0A1W4WV39 K0GG97 A0A2M4CIP8 A0A182N737 A0A0M4F6Q7 A0A0Q9X2S3 A0A026W561 A0A1I8MBT4 A0A0R1E150 A0A087ZTA6 A0A1I8PAR6 A0A0A1X4H1 A0A182VYR9 A0A1J1J616 A0A0N8P1E9 A0A2M3ZZK0 Q16N74 P13607-7 T1E1Y4 A0A0R1E1E2 A0A1J1J6V9 A0A0A1WY87 K0GLX7 A0A1Q3FV87 V9I6A9 K0GEZ1 A0A0Q9XCU5 A0A0Q9WBZ0 A0A1Q3FV30 A0A1Q3G4J5 K0GEX8 K0GEY8 A0A0K8UY58 A0A034W1V5 A0A1Q3FV07 A0A1B6MM45 A0A0Q9W0Q7
K0GG88 A0A2W1BK20 A0A2A4JV07 A0A2A4JUC5 A0A2H1VI27 K0GLY7 K0GG86 K0GEX7 K0G7C4 K0G7B9 A0A158NIT2 Q7PGN1 A0A1W4X4P7 K0GLY2 A0A182QT13 A0A182XW64 A0A1L8DEW0 A0A182PSN6 A0A3L8D359 V9I6B8 N6T2P0 A0A2A3E444 U4UIT1 E9IZS4 A0A158NIT1 E2AIZ4 F5HKJ1 F5HKJ2 A0A1Q3G4G9 A0A1Q3FUU5 A0A182LGE6 A0A0K8TRD8 A0A182IU13 F5HKJ4 A0A151IUE3 A0A1W4WV71 A0A0Q9X358 A0A0C9R0I4 A0A1L8DF50 A0A0Q9WBH9 Q27766 A0A1W4X5W5 A0A2M3ZZE0 A0A1W4WUY0 A0A1W4WV42 I5ANS6 A0A1Y0AWP8 A0A1W4X5V6 A0A2M4CSX9 A0A2M4CUW4 A0A182RQ21 A0A1W4X5S4 A0A2M4A020 A0A2M4A3F2 A0A1L8DEU4 A0A2M4CUE1 A0A1W4X5Q8 A0A1W4WV39 K0GG97 A0A2M4CIP8 A0A182N737 A0A0M4F6Q7 A0A0Q9X2S3 A0A026W561 A0A1I8MBT4 A0A0R1E150 A0A087ZTA6 A0A1I8PAR6 A0A0A1X4H1 A0A182VYR9 A0A1J1J616 A0A0N8P1E9 A0A2M3ZZK0 Q16N74 P13607-7 T1E1Y4 A0A0R1E1E2 A0A1J1J6V9 A0A0A1WY87 K0GLX7 A0A1Q3FV87 V9I6A9 K0GEZ1 A0A0Q9XCU5 A0A0Q9WBZ0 A0A1Q3FV30 A0A1Q3G4J5 K0GEX8 K0GEY8 A0A0K8UY58 A0A034W1V5 A0A1Q3FV07 A0A1B6MM45 A0A0Q9W0Q7
PDB
3WGV
E-value=0,
Score=3968
Ontologies
GO
GO:0046872
GO:0008556
GO:0016021
GO:0005524
GO:0006813
GO:0036376
GO:1990573
GO:0005391
GO:0030007
GO:0006883
GO:0005623
GO:0015991
GO:0005918
GO:0007630
GO:0005634
GO:0051124
GO:0001894
GO:0007268
GO:0016323
GO:0060439
GO:0008340
GO:0035158
GO:0009266
GO:0007605
GO:0009612
GO:0050905
GO:0019991
GO:0005886
GO:0008344
GO:0005890
GO:0006812
GO:0035159
GO:0005737
GO:0007626
GO:0000166
GO:0006814
GO:0006270
GO:0042555
GO:0015930
Topology
Subcellular location
Membrane
Cell membrane
Cell membrane
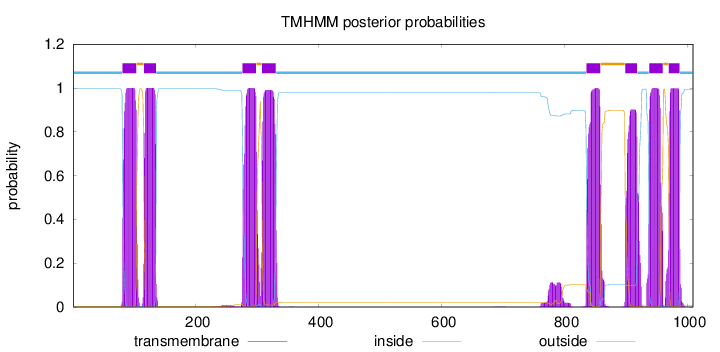
Length:
1009
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
172.12338
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99690
inside
1 - 80
TMhelix
81 - 103
outside
104 - 115
TMhelix
116 - 135
inside
136 - 275
TMhelix
276 - 298
outside
299 - 307
TMhelix
308 - 330
inside
331 - 835
TMhelix
836 - 858
outside
859 - 898
TMhelix
899 - 918
inside
919 - 937
TMhelix
938 - 960
outside
961 - 969
TMhelix
970 - 987
inside
988 - 1009
Population Genetic Test Statistics
Pi
241.608966
Theta
198.924669
Tajima's D
0.794025
CLR
0.392542
CSRT
0.604669766511674
Interpretation
Uncertain
