Gene
KWMTBOMO15313 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005142
Annotation
PREDICTED:_alpha-mannosidase_2_[Papilio_machaon]
Full name
Alpha-mannosidase
Location in the cell
Cytoplasmic Reliability : 1.352 Nuclear Reliability : 1.204
Sequence
CDS
ATGAGGATCAGACGTTTATCGGCGTTTTTTTGGATGACGTGTATTATAGCTTTTATTTTTATTTTATACGTGATGACAGACTTAAGTTTTAATTTACCTAGTGTTAAACCAGCGATAAACATAGATGAGAACAAGTGGTCTGCCTTTGAATCAAAATTACGTCATATTGAAAATGAACTTGATCAACATCATGCTGTTGTTGGTGAAATAAAATCAGCCATGAGCCAAATTATAGAACAGTCAAATGAATATCACCCGCCACAGAAAAGACCAAGGCAAATGGAGAAAAACAAAAAAATAAGAGAGTTCAAAGTAGTAGGAGATGAGCCACCAAGGATGCACATAAAATTGAATGAATCAACTTGTCCCATTCAGAAATACTCAGTTCCAAAAACTGATGTGCAGATGTTATCCTTGTATGAAAGGATTATGTTTGATGACAAAGATGGAGGTGTTTGGAAACAAGGATGGAATATAGAATACAATGAAAACCAATGGAGTCAGAAAAACAAATTGAAAGTTTTTATTGTACCACATTCACACAATGACCCAGGTTGGTTAAAAACCTTCGAAAATTATTACAAGTCACAAACAAGAGCAATATTCAATAATATGATAGAGAAATTGAACGAGGGTGTAGGGCGTAAGTTTATATGGGCCGAAATAAGTTTTCTATCGCTGTGGTGGAATAACGATGCTACTGAGAAAGACAAGACTGCTTTCCTGAATTTATTGAAATCAAAGAAACTTGAAATCGTGACAGGCGGATGGGTTATGAACGACGAGGCAAATTCACATTGGCTGTCTGTAGTGCAACAATTGACCACCGGACATCAGTGGCTACTCGACAATTTGGGATATGTACCCAAAAACGCATGGTCCATTGATCCGTTCGGTTATTCCAGCGCGCAACCCTACCTCCTCAAGATAGCCGGTTTCGATAATTCCATGATACAGAGGGTACACTATAGAATTAAGAAAGAATTGGCTTCGAATAGACAGTTGGAATTTAGATGGAGACAGTTATGGGATGGTGTTGGGAAAACTGATATGTTCACACATTTGTTCCCGTTTTATTCGTACGATGTTCCTCACTCGTGCGGTCCGGATCCTAAGATATGTTGTCAATTTGATTTCAAACGTCTACCCGGTAACGGGGTTACTTGTCCGTGGGGCGTTGCACCTCAGAGGATAACCCAGAAGAATGTGGATGAAAGAGCGTTCAAGATATTAGATCAGTGGAGGAAGAAGGCTCAACTCTACAGAAGTAACGTGTTGTTCTTCCCTCTCGGTGACGACTTCCGGTACGACCACGCCAACGAGTGGGACAATCAGTACCAGAACTACGACATGCTGATAGAGTACATCAACAGTAACGATGCTTGGAACACTGAGGTACAATACGGTACACTCGAAGATTACTTCAAAGCATTACACGATGAAGTGAAATTATCAAACTTTCCGGTTCTGTCCGGGGACTTTTTCACGTACGCCGATCGGAACCAGCACTACTGGAGCGGGTACTACACCTCCAGACCGTTTTATAAGAACATGGATAGAGTTTTACTGGCCTATGTCAGGGCAGCCGAAATAATAACAGCTCAAGCGACGGACACGCATGCTGTGTCGTACATAATGTCGCTTCAGCTGAGGGACCGGGTCGAGCAGGCCCGCAGGTCGCTCTCCCTGTTCCAGCATCACGACGGCATCACCGGCACCTCCAGGGACGAGGTCAGGGAGGACTACGCCAAGAAGATGCTAACAGCAATAAAATACAGTCAATCCGCCATCCAACAAGCTGCCTACTACTTGCTGAAGCAGCCGCATATACAGGATCAAACGCAAGAGGACATATTCCTCGATGTGGACGACATCTGGCGGCGGCACGACGAAATACCCTCCAGGATCACGATTGCCTTGGACGTGGTGTCTCCTTCCAGGCGCCTCGTGCTTTACAACGCGCTGGCGTTCAGAAGACAAGAAATATTAACAGTGCTAGTGTCAACCCCGCACGTTGAAGTTAGTAAGAAACCAATATCGGGGTATTAA
Protein
MRIRRLSAFFWMTCIIAFIFILYVMTDLSFNLPSVKPAINIDENKWSAFESKLRHIENELDQHHAVVGEIKSAMSQIIEQSNEYHPPQKRPRQMEKNKKIREFKVVGDEPPRMHIKLNESTCPIQKYSVPKTDVQMLSLYERIMFDDKDGGVWKQGWNIEYNENQWSQKNKLKVFIVPHSHNDPGWLKTFENYYKSQTRAIFNNMIEKLNEGVGRKFIWAEISFLSLWWNNDATEKDKTAFLNLLKSKKLEIVTGGWVMNDEANSHWLSVVQQLTTGHQWLLDNLGYVPKNAWSIDPFGYSSAQPYLLKIAGFDNSMIQRVHYRIKKELASNRQLEFRWRQLWDGVGKTDMFTHLFPFYSYDVPHSCGPDPKICCQFDFKRLPGNGVTCPWGVAPQRITQKNVDERAFKILDQWRKKAQLYRSNVLFFPLGDDFRYDHANEWDNQYQNYDMLIEYINSNDAWNTEVQYGTLEDYFKALHDEVKLSNFPVLSGDFFTYADRNQHYWSGYYTSRPFYKNMDRVLLAYVRAAEIITAQATDTHAVSYIMSLQLRDRVEQARRSLSLFQHHDGITGTSRDEVREDYAKKMLTAIKYSQSAIQQAAYYLLKQPHIQDQTQEDIFLDVDDIWRRHDEIPSRITIALDVVSPSRRLVLYNALAFRRQEILTVLVSTPHVEVSKKPISGY
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the glycosyl hydrolase 38 family.
Uniprot
A0A2A4JW66
A0A2W1BFQ4
A0A194RKL8
A0A194PGJ2
A0A212F8Y0
H9J6K0
+ More
A0A067R639 D6WTA9 A0A1Y1L0D7 V5GX04 N6TR79 A0A2P8Z8I7 A0A026WXN3 A0A195DUT2 A0A151I6B5 A0A158NSN6 A0A0N0BDG4 A0A195CCN0 A0A0L7R1D6 A0A195EXK8 A0A151WJA6 A0A1B6LSU4 E2C820 E2AKY7 A0A0C9R4X5 A0A087ZZQ3 A0A2A3EQ80 A0A232FIH3 A0A1B6CBG0 K7J1I1 A0A0C9RFU8 E9IUM4 A0A023F1Z6 A0A0P4VK91 A0A224X5V1 A0A2M3ZE22 A0A182IRW0 A0A0V0G6A5 A0A0P4ZKB6 A0A0P5LI91 A0A0P6BXP3 A0A0P4ZQ30 A0A0P5CYJ7 A0A0P6IIZ0 A0A0P5DBH3 A0A0P4ZIN8 A0A0P5J864 A0A2M4BAW1 A0A0P6EAM1 A0A0P5EAM2 A0A0P5DAP0 A0A0P5DW21 A0A0P4ZSN2 A0A0P5HTF8 A0A0P5D5J5 A0A2M4BBI3 A0A0P5JH22 A0A0P5D6W5 A0A0P5S8D1 A0A0P5CLK9 A0A0N8AE08 A0A0P5AKD0 A0A0P5AKF7 A0A0P5CQT8 A0A0P5UDR8 A0A0P5J1W0 A0A0P5IWE5 E0VV82 T1HHY4 A0A2M4AC47 A0A2M4ACL9 A0A084VTM1 A0A0J7KHF7 W5JK63 A0A182P424 U5EZM2 A0A182JSW1 A0A182GXK5 A0A182FPQ5 A0A182YG16 A0A182I875 A0A182KML8 Q7PMM6 A0A1Z5LHE7 A0A2R5LA05 Q16NG2 A0A1Q3FDG5 F4W8A6 A0A182UAA6 A0A182V343 B0WS34 A0A1Y9J179 A0A1W4XMZ0 A0A1W4XNT9 A0A0P5A1B3 A0A1Y9HF01 A0A182VWZ0 A0A1B0CQQ4 A0A182NDY6
A0A067R639 D6WTA9 A0A1Y1L0D7 V5GX04 N6TR79 A0A2P8Z8I7 A0A026WXN3 A0A195DUT2 A0A151I6B5 A0A158NSN6 A0A0N0BDG4 A0A195CCN0 A0A0L7R1D6 A0A195EXK8 A0A151WJA6 A0A1B6LSU4 E2C820 E2AKY7 A0A0C9R4X5 A0A087ZZQ3 A0A2A3EQ80 A0A232FIH3 A0A1B6CBG0 K7J1I1 A0A0C9RFU8 E9IUM4 A0A023F1Z6 A0A0P4VK91 A0A224X5V1 A0A2M3ZE22 A0A182IRW0 A0A0V0G6A5 A0A0P4ZKB6 A0A0P5LI91 A0A0P6BXP3 A0A0P4ZQ30 A0A0P5CYJ7 A0A0P6IIZ0 A0A0P5DBH3 A0A0P4ZIN8 A0A0P5J864 A0A2M4BAW1 A0A0P6EAM1 A0A0P5EAM2 A0A0P5DAP0 A0A0P5DW21 A0A0P4ZSN2 A0A0P5HTF8 A0A0P5D5J5 A0A2M4BBI3 A0A0P5JH22 A0A0P5D6W5 A0A0P5S8D1 A0A0P5CLK9 A0A0N8AE08 A0A0P5AKD0 A0A0P5AKF7 A0A0P5CQT8 A0A0P5UDR8 A0A0P5J1W0 A0A0P5IWE5 E0VV82 T1HHY4 A0A2M4AC47 A0A2M4ACL9 A0A084VTM1 A0A0J7KHF7 W5JK63 A0A182P424 U5EZM2 A0A182JSW1 A0A182GXK5 A0A182FPQ5 A0A182YG16 A0A182I875 A0A182KML8 Q7PMM6 A0A1Z5LHE7 A0A2R5LA05 Q16NG2 A0A1Q3FDG5 F4W8A6 A0A182UAA6 A0A182V343 B0WS34 A0A1Y9J179 A0A1W4XMZ0 A0A1W4XNT9 A0A0P5A1B3 A0A1Y9HF01 A0A182VWZ0 A0A1B0CQQ4 A0A182NDY6
EC Number
3.2.1.-
Pubmed
EMBL
NWSH01000562
PCG75640.1
KZ150095
PZC73618.1
KQ460045
KPJ18082.1
+ More
KQ459604 KPI92422.1 AGBW02009658 OWR50206.1 BABH01019831 KK852934 KDR13614.1 KQ971352 EFA06316.1 GEZM01071358 JAV65870.1 GALX01002174 JAB66292.1 APGK01057855 APGK01057856 APGK01057857 KB741282 ENN70826.1 PYGN01000147 PSN52814.1 KK107078 QOIP01000011 EZA60501.1 RLU16846.1 KQ980322 KYN16606.1 KQ976400 KYM92668.1 ADTU01025073 ADTU01025074 ADTU01025075 KQ435863 KOX70418.1 KQ978023 KYM97981.1 KQ414667 KOC64685.1 KQ981948 KYN32614.1 KQ983043 KYQ47914.1 GEBQ01013209 JAT26768.1 GL453506 EFN75938.1 GL440425 EFN65904.1 GBYB01005913 GBYB01011354 GBYB01013488 GBYB01013723 JAG75680.1 JAG81121.1 JAG83255.1 JAG83490.1 KZ288194 PBC33888.1 NNAY01000155 OXU30485.1 GEDC01026500 JAS10798.1 GBYB01005906 JAG75673.1 GL765999 EFZ15726.1 GBBI01003636 JAC15076.1 GDKW01003685 JAI52910.1 GFTR01008561 JAW07865.1 GGFM01006002 MBW26753.1 GECL01002524 JAP03600.1 GDIP01211271 JAJ12131.1 GDIQ01169247 JAK82478.1 GDIP01008771 JAM94944.1 GDIP01211270 JAJ12132.1 GDIP01163766 JAJ59636.1 GDIQ01005472 JAN89265.1 GDIP01163767 JAJ59635.1 GDIP01212278 JAJ11124.1 GDIQ01203735 JAK47990.1 GGFJ01001026 MBW50167.1 GDIQ01145858 GDIQ01065542 JAN29195.1 GDIP01162821 JAJ60581.1 GDIP01162820 JAJ60582.1 GDIP01156672 JAJ66730.1 GDIP01212279 JAJ11123.1 GDIQ01246032 JAK05693.1 GDIP01160925 JAJ62477.1 GGFJ01001027 MBW50168.1 GDIQ01252080 GDIQ01250680 GDIQ01247419 GDIQ01205391 GDIQ01202240 GDIQ01099829 GDIQ01098480 GDIQ01086140 JAK46334.1 GDIP01176215 GDIP01141337 JAJ47187.1 GDIQ01252081 GDIQ01250679 GDIQ01247420 GDIQ01205390 GDIQ01203734 GDIQ01202241 GDIQ01187751 GDIQ01099830 GDIQ01098479 GDIQ01096821 JAL51896.1 GDIP01168720 JAJ54682.1 GDIP01156673 JAJ66729.1 GDIP01198078 JAJ25324.1 GDIP01198077 JAJ25325.1 GDIP01168070 JAJ55332.1 GDIP01131684 LRGB01000031 JAL72030.1 KZS21240.1 GDIQ01208140 JAK43585.1 GDIQ01208141 JAK43584.1 DS235804 EEB17288.1 ACPB03005842 GGFK01005036 MBW38357.1 GGFK01005027 MBW38348.1 ATLV01016361 KE525079 KFB41315.1 LBMM01007530 KMQ89646.1 ADMH02001205 ETN63703.1 GANO01000532 JAB59339.1 JXUM01095509 JXUM01095510 KQ564200 KXJ72595.1 APCN01002661 AAAB01008978 EAA13579.5 GFJQ02000144 JAW06826.1 GGLE01002206 MBY06332.1 CH477824 EAT35901.1 GFDL01009486 JAV25559.1 GL887898 EGI69601.1 DS232064 EDS33653.1 GDIP01205412 JAJ17990.1 AJWK01023876
KQ459604 KPI92422.1 AGBW02009658 OWR50206.1 BABH01019831 KK852934 KDR13614.1 KQ971352 EFA06316.1 GEZM01071358 JAV65870.1 GALX01002174 JAB66292.1 APGK01057855 APGK01057856 APGK01057857 KB741282 ENN70826.1 PYGN01000147 PSN52814.1 KK107078 QOIP01000011 EZA60501.1 RLU16846.1 KQ980322 KYN16606.1 KQ976400 KYM92668.1 ADTU01025073 ADTU01025074 ADTU01025075 KQ435863 KOX70418.1 KQ978023 KYM97981.1 KQ414667 KOC64685.1 KQ981948 KYN32614.1 KQ983043 KYQ47914.1 GEBQ01013209 JAT26768.1 GL453506 EFN75938.1 GL440425 EFN65904.1 GBYB01005913 GBYB01011354 GBYB01013488 GBYB01013723 JAG75680.1 JAG81121.1 JAG83255.1 JAG83490.1 KZ288194 PBC33888.1 NNAY01000155 OXU30485.1 GEDC01026500 JAS10798.1 GBYB01005906 JAG75673.1 GL765999 EFZ15726.1 GBBI01003636 JAC15076.1 GDKW01003685 JAI52910.1 GFTR01008561 JAW07865.1 GGFM01006002 MBW26753.1 GECL01002524 JAP03600.1 GDIP01211271 JAJ12131.1 GDIQ01169247 JAK82478.1 GDIP01008771 JAM94944.1 GDIP01211270 JAJ12132.1 GDIP01163766 JAJ59636.1 GDIQ01005472 JAN89265.1 GDIP01163767 JAJ59635.1 GDIP01212278 JAJ11124.1 GDIQ01203735 JAK47990.1 GGFJ01001026 MBW50167.1 GDIQ01145858 GDIQ01065542 JAN29195.1 GDIP01162821 JAJ60581.1 GDIP01162820 JAJ60582.1 GDIP01156672 JAJ66730.1 GDIP01212279 JAJ11123.1 GDIQ01246032 JAK05693.1 GDIP01160925 JAJ62477.1 GGFJ01001027 MBW50168.1 GDIQ01252080 GDIQ01250680 GDIQ01247419 GDIQ01205391 GDIQ01202240 GDIQ01099829 GDIQ01098480 GDIQ01086140 JAK46334.1 GDIP01176215 GDIP01141337 JAJ47187.1 GDIQ01252081 GDIQ01250679 GDIQ01247420 GDIQ01205390 GDIQ01203734 GDIQ01202241 GDIQ01187751 GDIQ01099830 GDIQ01098479 GDIQ01096821 JAL51896.1 GDIP01168720 JAJ54682.1 GDIP01156673 JAJ66729.1 GDIP01198078 JAJ25324.1 GDIP01198077 JAJ25325.1 GDIP01168070 JAJ55332.1 GDIP01131684 LRGB01000031 JAL72030.1 KZS21240.1 GDIQ01208140 JAK43585.1 GDIQ01208141 JAK43584.1 DS235804 EEB17288.1 ACPB03005842 GGFK01005036 MBW38357.1 GGFK01005027 MBW38348.1 ATLV01016361 KE525079 KFB41315.1 LBMM01007530 KMQ89646.1 ADMH02001205 ETN63703.1 GANO01000532 JAB59339.1 JXUM01095509 JXUM01095510 KQ564200 KXJ72595.1 APCN01002661 AAAB01008978 EAA13579.5 GFJQ02000144 JAW06826.1 GGLE01002206 MBY06332.1 CH477824 EAT35901.1 GFDL01009486 JAV25559.1 GL887898 EGI69601.1 DS232064 EDS33653.1 GDIP01205412 JAJ17990.1 AJWK01023876
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000005204
UP000027135
+ More
UP000007266 UP000019118 UP000245037 UP000053097 UP000279307 UP000078492 UP000078540 UP000005205 UP000053105 UP000078542 UP000053825 UP000078541 UP000075809 UP000008237 UP000000311 UP000005203 UP000242457 UP000215335 UP000002358 UP000075880 UP000076858 UP000009046 UP000015103 UP000030765 UP000036403 UP000000673 UP000075885 UP000075881 UP000069940 UP000249989 UP000069272 UP000076408 UP000075840 UP000075882 UP000007062 UP000008820 UP000007755 UP000075902 UP000075903 UP000002320 UP000076407 UP000192223 UP000075900 UP000075920 UP000092461 UP000075884
UP000007266 UP000019118 UP000245037 UP000053097 UP000279307 UP000078492 UP000078540 UP000005205 UP000053105 UP000078542 UP000053825 UP000078541 UP000075809 UP000008237 UP000000311 UP000005203 UP000242457 UP000215335 UP000002358 UP000075880 UP000076858 UP000009046 UP000015103 UP000030765 UP000036403 UP000000673 UP000075885 UP000075881 UP000069940 UP000249989 UP000069272 UP000076408 UP000075840 UP000075882 UP000007062 UP000008820 UP000007755 UP000075902 UP000075903 UP000002320 UP000076407 UP000192223 UP000075900 UP000075920 UP000092461 UP000075884
PRIDE
Pfam
Interpro
IPR011330
Glyco_hydro/deAcase_b/a-brl
+ More
IPR000602 Glyco_hydro_38_N
IPR015341 Glyco_hydro_38_cen
IPR011013 Gal_mutarotase_sf_dom
IPR027291 Glyco_hydro_38_N_sf
IPR037094 Glyco_hydro_38_cen_sf
IPR011682 Glyco_hydro_38_C
IPR013780 Glyco_hydro_b
IPR028995 Glyco_hydro_57/38_cen_sf
IPR041566 AManII_C
IPR003347 JmjC_dom
IPR001320 Iontro_rcpt
IPR041667 Cupin_8
IPR038765 Papain-like_cys_pep_sf
IPR005078 Peptidase_C54
IPR000602 Glyco_hydro_38_N
IPR015341 Glyco_hydro_38_cen
IPR011013 Gal_mutarotase_sf_dom
IPR027291 Glyco_hydro_38_N_sf
IPR037094 Glyco_hydro_38_cen_sf
IPR011682 Glyco_hydro_38_C
IPR013780 Glyco_hydro_b
IPR028995 Glyco_hydro_57/38_cen_sf
IPR041566 AManII_C
IPR003347 JmjC_dom
IPR001320 Iontro_rcpt
IPR041667 Cupin_8
IPR038765 Papain-like_cys_pep_sf
IPR005078 Peptidase_C54
Gene 3D
ProteinModelPortal
A0A2A4JW66
A0A2W1BFQ4
A0A194RKL8
A0A194PGJ2
A0A212F8Y0
H9J6K0
+ More
A0A067R639 D6WTA9 A0A1Y1L0D7 V5GX04 N6TR79 A0A2P8Z8I7 A0A026WXN3 A0A195DUT2 A0A151I6B5 A0A158NSN6 A0A0N0BDG4 A0A195CCN0 A0A0L7R1D6 A0A195EXK8 A0A151WJA6 A0A1B6LSU4 E2C820 E2AKY7 A0A0C9R4X5 A0A087ZZQ3 A0A2A3EQ80 A0A232FIH3 A0A1B6CBG0 K7J1I1 A0A0C9RFU8 E9IUM4 A0A023F1Z6 A0A0P4VK91 A0A224X5V1 A0A2M3ZE22 A0A182IRW0 A0A0V0G6A5 A0A0P4ZKB6 A0A0P5LI91 A0A0P6BXP3 A0A0P4ZQ30 A0A0P5CYJ7 A0A0P6IIZ0 A0A0P5DBH3 A0A0P4ZIN8 A0A0P5J864 A0A2M4BAW1 A0A0P6EAM1 A0A0P5EAM2 A0A0P5DAP0 A0A0P5DW21 A0A0P4ZSN2 A0A0P5HTF8 A0A0P5D5J5 A0A2M4BBI3 A0A0P5JH22 A0A0P5D6W5 A0A0P5S8D1 A0A0P5CLK9 A0A0N8AE08 A0A0P5AKD0 A0A0P5AKF7 A0A0P5CQT8 A0A0P5UDR8 A0A0P5J1W0 A0A0P5IWE5 E0VV82 T1HHY4 A0A2M4AC47 A0A2M4ACL9 A0A084VTM1 A0A0J7KHF7 W5JK63 A0A182P424 U5EZM2 A0A182JSW1 A0A182GXK5 A0A182FPQ5 A0A182YG16 A0A182I875 A0A182KML8 Q7PMM6 A0A1Z5LHE7 A0A2R5LA05 Q16NG2 A0A1Q3FDG5 F4W8A6 A0A182UAA6 A0A182V343 B0WS34 A0A1Y9J179 A0A1W4XMZ0 A0A1W4XNT9 A0A0P5A1B3 A0A1Y9HF01 A0A182VWZ0 A0A1B0CQQ4 A0A182NDY6
A0A067R639 D6WTA9 A0A1Y1L0D7 V5GX04 N6TR79 A0A2P8Z8I7 A0A026WXN3 A0A195DUT2 A0A151I6B5 A0A158NSN6 A0A0N0BDG4 A0A195CCN0 A0A0L7R1D6 A0A195EXK8 A0A151WJA6 A0A1B6LSU4 E2C820 E2AKY7 A0A0C9R4X5 A0A087ZZQ3 A0A2A3EQ80 A0A232FIH3 A0A1B6CBG0 K7J1I1 A0A0C9RFU8 E9IUM4 A0A023F1Z6 A0A0P4VK91 A0A224X5V1 A0A2M3ZE22 A0A182IRW0 A0A0V0G6A5 A0A0P4ZKB6 A0A0P5LI91 A0A0P6BXP3 A0A0P4ZQ30 A0A0P5CYJ7 A0A0P6IIZ0 A0A0P5DBH3 A0A0P4ZIN8 A0A0P5J864 A0A2M4BAW1 A0A0P6EAM1 A0A0P5EAM2 A0A0P5DAP0 A0A0P5DW21 A0A0P4ZSN2 A0A0P5HTF8 A0A0P5D5J5 A0A2M4BBI3 A0A0P5JH22 A0A0P5D6W5 A0A0P5S8D1 A0A0P5CLK9 A0A0N8AE08 A0A0P5AKD0 A0A0P5AKF7 A0A0P5CQT8 A0A0P5UDR8 A0A0P5J1W0 A0A0P5IWE5 E0VV82 T1HHY4 A0A2M4AC47 A0A2M4ACL9 A0A084VTM1 A0A0J7KHF7 W5JK63 A0A182P424 U5EZM2 A0A182JSW1 A0A182GXK5 A0A182FPQ5 A0A182YG16 A0A182I875 A0A182KML8 Q7PMM6 A0A1Z5LHE7 A0A2R5LA05 Q16NG2 A0A1Q3FDG5 F4W8A6 A0A182UAA6 A0A182V343 B0WS34 A0A1Y9J179 A0A1W4XMZ0 A0A1W4XNT9 A0A0P5A1B3 A0A1Y9HF01 A0A182VWZ0 A0A1B0CQQ4 A0A182NDY6
PDB
3EJU
E-value=4.7361e-171,
Score=1545
Ontologies
PATHWAY
GO
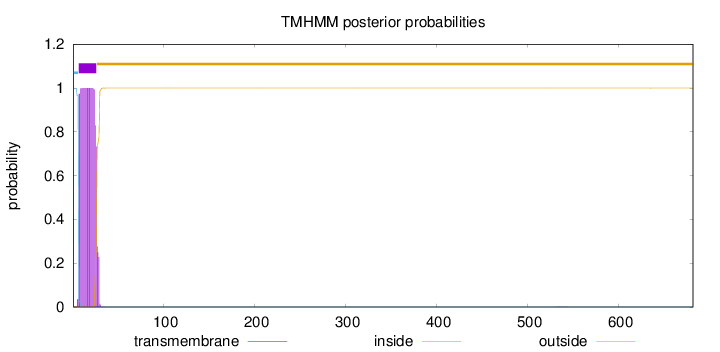
Topology
Length:
682
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.35005
Exp number, first 60 AAs:
20.34669
Total prob of N-in:
0.99993
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 682
Population Genetic Test Statistics
Pi
231.484862
Theta
194.695063
Tajima's D
1.102676
CLR
0.000516
CSRT
0.689815509224539
Interpretation
Uncertain
