Pre Gene Modal
BGIBMGA005142
Annotation
PREDICTED:_alpha-mannosidase_2_[Papilio_xuthus]
Full name
Alpha-mannosidase
Location in the cell
Extracellular Reliability : 1.005 Nuclear Reliability : 1.054
Sequence
CDS
ATGAACGGGCTCGTCAAAACCATTGTGTCGAGGGACGGAGTGGCCACACCCGTGCACATGGACTTCGTGCAGTACGACACGCAGAAGGGGCGGGACAACAACAGCGGAGCCTACCTGTTCATACCGAGCGGGCCCGCCAAGGACTTCAAGTCGGATCCGTACCCCGAGATCGTGGTCACCGAAGGCATACACAAGTCCACGCTCTACGCCGCCCTCGTCGGACCCAAAACCGCAGAAATTATACTCTCTGTGACGGTGTACAACAACCCGTCGCTTCCGCACGCCGAGCTGGAGGTCAGCAGCGCGCTGCTCATCGACCCGCAGGTGGACGACCTGGAGCTGGCGCTGCGGCTATCCACTAGCGTCCGGAACGGAGACGTCTTCTACACAGACTTAAATGGGATGCAGATGATAAAACGTCGTTACTTCGAGAAGATTCCGCTGCAGGCGAACTTTTATCCGCTGCCCGCCGCCGCCTACATCGAGGACGAGAACATCAGGTTCACGGTGCTCACTTCCACTCCGTTAGGAATGGCTGCCTTACAGCCGGGACAAATTGAGGTGATGCAAGACAGGCGCCTGAGCAGAGACGACAACCGAGGCATGAACCAAGGCGTGTTGGACAACGTGAGGACGCGCCACACATTCAGGCTCATAGCGGAGCCGGCCCTCGGGCACTGCGCCAAGCCGCCGTCTGATCACCCAAGCGGGTGGCTGACGCTGGGTAGTCACGTGTCGCAGCAGATGCTCCTGCAGCCGCCGCTCGTGCTGCACTACACGCCGCGCGGAGACGAGCCCCGCCCCAACTACCAGAGGGCCTCCGTCAACGCCGCCGACATCGTGCTGGCCTCCTACAGAACCCACACCACCACTAAGAAGGAAGGCACGAATTTACACGGGATGGGGCTGACGTTCCAAAGGGTAGCCCTCGACACGTGCTACGGTCACAAGGACACCGTCGAAGCGTATCCGCTAAGTGACGGACAGTTCCGTCTGTCAGATTTGATAGACGTGAAATCAGATCGTGTTTACGAAAGCACGTTGACTTTCAATCAGATAGACAAGCAAGTGTTAGACGGAATCATCACGCTGTGCCCCATGGAGATCCGATCCGTGTTCGTCAATCAAACTATAAGGAACGGATGA
Protein
MNGLVKTIVSRDGVATPVHMDFVQYDTQKGRDNNSGAYLFIPSGPAKDFKSDPYPEIVVTEGIHKSTLYAALVGPKTAEIILSVTVYNNPSLPHAELEVSSALLIDPQVDDLELALRLSTSVRNGDVFYTDLNGMQMIKRRYFEKIPLQANFYPLPAAAYIEDENIRFTVLTSTPLGMAALQPGQIEVMQDRRLSRDDNRGMNQGVLDNVRTRHTFRLIAEPALGHCAKPPSDHPSGWLTLGSHVSQQMLLQPPLVLHYTPRGDEPRPNYQRASVNAADIVLASYRTHTTTKKEGTNLHGMGLTFQRVALDTCYGHKDTVEAYPLSDGQFRLSDLIDVKSDRVYESTLTFNQIDKQVLDGIITLCPMEIRSVFVNQTIRNG
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the glycosyl hydrolase 38 family.
Uniprot
H9J6K0
A0A2H1V2N4
A0A2W1BFQ4
A0A194PGJ2
A0A2A4JW66
A0A194RKL8
+ More
A0A212F8Y0 A0A0L7LNU1 A0A2H1W4P3 U4UDM8 N6TR79 A0A2J7QCV6 V5H0D6 A0A067R639 K7J1I1 A0A2R7WF52 A0A232FIH3 A0A1Y1L0D7 D6WTA9 A0A0L7R1D6 E2C820 A0A3Q0ILT9 A0A0V0G6A5 E9IUM4 A0A0N0BDG4 A0A1B0GEU5 A0A195DUT2 A0A023F1Z6 A0A1A9VSM3 F4W8A6 A0A151WJA6 A0A154PIT6 A0A1A9ZM93 A0A195CCN0 A0A158NSN6 A0A151I6B5 A0A0C9R4X5 A0A2A3EQ80 A0A087ZZQ3 A0A0T6B8D9 A0A195EXK8 A0A1B0AVB9 A0A1A9YBN7 A0A026WXN3 A0A0Q9WPC2 A0A0Q9WQX2 A0A224X5V1 E2AKY7 B4LW20 A0A0P4VK91 A0A1B6CBG0 B4GP61 A0A0A9XUQ5 Q29BM2 A0A3B0K5A5 A0A146M558 B4JEK9 A0A1I8PWA3 A0A034VRR0 A0A023EW91 B4K4Z8 T1HHY4 B0WS34 A0A0N4UP28 A0A0K8V1K2 A0A1B6LSU4 B3NZS4 A0A1I8MUB4 A0A182GXK5 A0A0K8UCQ8 U5EZM2 A0A1B6IHW2 A0A2S2Q527 A0A0A1X4Y7 A0A1I8CJ16 A0A0L0C6C3 A0A1Q3FDG5 A0A1L8E1R2 Q16NG2 A0A1L8DR70 J9K9U5 A0A2S2P0E5 W8C710 W8BNH3 E0VV82 A0A2S2PN40 A0A1B6FR71
A0A212F8Y0 A0A0L7LNU1 A0A2H1W4P3 U4UDM8 N6TR79 A0A2J7QCV6 V5H0D6 A0A067R639 K7J1I1 A0A2R7WF52 A0A232FIH3 A0A1Y1L0D7 D6WTA9 A0A0L7R1D6 E2C820 A0A3Q0ILT9 A0A0V0G6A5 E9IUM4 A0A0N0BDG4 A0A1B0GEU5 A0A195DUT2 A0A023F1Z6 A0A1A9VSM3 F4W8A6 A0A151WJA6 A0A154PIT6 A0A1A9ZM93 A0A195CCN0 A0A158NSN6 A0A151I6B5 A0A0C9R4X5 A0A2A3EQ80 A0A087ZZQ3 A0A0T6B8D9 A0A195EXK8 A0A1B0AVB9 A0A1A9YBN7 A0A026WXN3 A0A0Q9WPC2 A0A0Q9WQX2 A0A224X5V1 E2AKY7 B4LW20 A0A0P4VK91 A0A1B6CBG0 B4GP61 A0A0A9XUQ5 Q29BM2 A0A3B0K5A5 A0A146M558 B4JEK9 A0A1I8PWA3 A0A034VRR0 A0A023EW91 B4K4Z8 T1HHY4 B0WS34 A0A0N4UP28 A0A0K8V1K2 A0A1B6LSU4 B3NZS4 A0A1I8MUB4 A0A182GXK5 A0A0K8UCQ8 U5EZM2 A0A1B6IHW2 A0A2S2Q527 A0A0A1X4Y7 A0A1I8CJ16 A0A0L0C6C3 A0A1Q3FDG5 A0A1L8E1R2 Q16NG2 A0A1L8DR70 J9K9U5 A0A2S2P0E5 W8C710 W8BNH3 E0VV82 A0A2S2PN40 A0A1B6FR71
EC Number
3.2.1.-
Pubmed
19121390
28756777
26354079
22118469
26227816
23537049
+ More
24845553 20075255 28648823 28004739 18362917 19820115 20798317 21282665 25474469 21719571 21347285 24508170 30249741 17994087 27129103 25401762 15632085 26823975 25348373 24945155 25315136 26483478 25830018 26829753 26108605 17510324 24495485 20566863
24845553 20075255 28648823 28004739 18362917 19820115 20798317 21282665 25474469 21719571 21347285 24508170 30249741 17994087 27129103 25401762 15632085 26823975 25348373 24945155 25315136 26483478 25830018 26829753 26108605 17510324 24495485 20566863
EMBL
BABH01019831
ODYU01000395
SOQ35107.1
KZ150095
PZC73618.1
KQ459604
+ More
KPI92422.1 NWSH01000562 PCG75640.1 KQ460045 KPJ18082.1 AGBW02009658 OWR50206.1 JTDY01000470 KOB77029.1 ODYU01006317 SOQ48060.1 KB632233 ERL90403.1 APGK01057855 APGK01057856 APGK01057857 KB741282 ENN70826.1 NEVH01015858 PNF26420.1 GALX01002173 JAB66293.1 KK852934 KDR13614.1 KK854722 PTY18168.1 NNAY01000155 OXU30485.1 GEZM01071358 JAV65870.1 KQ971352 EFA06316.1 KQ414667 KOC64685.1 GL453506 EFN75938.1 GECL01002524 JAP03600.1 GL765999 EFZ15726.1 KQ435863 KOX70418.1 CCAG010012590 KQ980322 KYN16606.1 GBBI01003636 JAC15076.1 GL887898 EGI69601.1 KQ983043 KYQ47914.1 KQ434905 KZC11228.1 KQ978023 KYM97981.1 ADTU01025073 ADTU01025074 ADTU01025075 KQ976400 KYM92668.1 GBYB01005913 GBYB01011354 GBYB01013488 GBYB01013723 JAG75680.1 JAG81121.1 JAG83255.1 JAG83490.1 KZ288194 PBC33888.1 LJIG01009178 KRT83600.1 KQ981948 KYN32614.1 JXJN01004205 KK107078 QOIP01000011 EZA60501.1 RLU16846.1 CH940650 KRF82740.1 KRF82741.1 GFTR01008561 JAW07865.1 GL440425 EFN65904.1 EDW66525.1 GDKW01003685 JAI52910.1 GEDC01026500 JAS10798.1 CH479186 EDW38944.1 GBHO01019052 JAG24552.1 CM000070 EAL26975.2 OUUW01000005 SPP81179.1 GDHC01004722 JAQ13907.1 CH916369 EDV93140.1 GAKP01014437 JAC44515.1 GAPW01000123 JAC13475.1 CH933806 EDW13969.1 ACPB03005842 DS232064 EDS33653.1 UYYG01000123 VDN53345.1 GDHF01019517 JAI32797.1 GEBQ01013209 JAT26768.1 CH954181 EDV49784.1 JXUM01095509 JXUM01095510 KQ564200 KXJ72595.1 GDHF01027895 JAI24419.1 GANO01000532 JAB59339.1 GECU01021213 JAS86493.1 GGMS01003497 MBY72700.1 GBXI01007913 JAD06379.1 JRES01000835 KNC27938.1 GFDL01009486 JAV25559.1 GFDF01001436 JAV12648.1 CH477824 EAT35901.1 GFDF01005259 JAV08825.1 ABLF02034554 GGMR01010298 MBY22917.1 GAMC01003919 JAC02637.1 GAMC01003920 JAC02636.1 DS235804 EEB17288.1 GGMR01018214 MBY30833.1 GECZ01017062 JAS52707.1
KPI92422.1 NWSH01000562 PCG75640.1 KQ460045 KPJ18082.1 AGBW02009658 OWR50206.1 JTDY01000470 KOB77029.1 ODYU01006317 SOQ48060.1 KB632233 ERL90403.1 APGK01057855 APGK01057856 APGK01057857 KB741282 ENN70826.1 NEVH01015858 PNF26420.1 GALX01002173 JAB66293.1 KK852934 KDR13614.1 KK854722 PTY18168.1 NNAY01000155 OXU30485.1 GEZM01071358 JAV65870.1 KQ971352 EFA06316.1 KQ414667 KOC64685.1 GL453506 EFN75938.1 GECL01002524 JAP03600.1 GL765999 EFZ15726.1 KQ435863 KOX70418.1 CCAG010012590 KQ980322 KYN16606.1 GBBI01003636 JAC15076.1 GL887898 EGI69601.1 KQ983043 KYQ47914.1 KQ434905 KZC11228.1 KQ978023 KYM97981.1 ADTU01025073 ADTU01025074 ADTU01025075 KQ976400 KYM92668.1 GBYB01005913 GBYB01011354 GBYB01013488 GBYB01013723 JAG75680.1 JAG81121.1 JAG83255.1 JAG83490.1 KZ288194 PBC33888.1 LJIG01009178 KRT83600.1 KQ981948 KYN32614.1 JXJN01004205 KK107078 QOIP01000011 EZA60501.1 RLU16846.1 CH940650 KRF82740.1 KRF82741.1 GFTR01008561 JAW07865.1 GL440425 EFN65904.1 EDW66525.1 GDKW01003685 JAI52910.1 GEDC01026500 JAS10798.1 CH479186 EDW38944.1 GBHO01019052 JAG24552.1 CM000070 EAL26975.2 OUUW01000005 SPP81179.1 GDHC01004722 JAQ13907.1 CH916369 EDV93140.1 GAKP01014437 JAC44515.1 GAPW01000123 JAC13475.1 CH933806 EDW13969.1 ACPB03005842 DS232064 EDS33653.1 UYYG01000123 VDN53345.1 GDHF01019517 JAI32797.1 GEBQ01013209 JAT26768.1 CH954181 EDV49784.1 JXUM01095509 JXUM01095510 KQ564200 KXJ72595.1 GDHF01027895 JAI24419.1 GANO01000532 JAB59339.1 GECU01021213 JAS86493.1 GGMS01003497 MBY72700.1 GBXI01007913 JAD06379.1 JRES01000835 KNC27938.1 GFDL01009486 JAV25559.1 GFDF01001436 JAV12648.1 CH477824 EAT35901.1 GFDF01005259 JAV08825.1 ABLF02034554 GGMR01010298 MBY22917.1 GAMC01003919 JAC02637.1 GAMC01003920 JAC02636.1 DS235804 EEB17288.1 GGMR01018214 MBY30833.1 GECZ01017062 JAS52707.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000037510
+ More
UP000030742 UP000019118 UP000235965 UP000027135 UP000002358 UP000215335 UP000007266 UP000053825 UP000008237 UP000079169 UP000053105 UP000092444 UP000078492 UP000078200 UP000007755 UP000075809 UP000076502 UP000092445 UP000078542 UP000005205 UP000078540 UP000242457 UP000005203 UP000078541 UP000092460 UP000092443 UP000053097 UP000279307 UP000008792 UP000000311 UP000008744 UP000001819 UP000268350 UP000001070 UP000095300 UP000009192 UP000015103 UP000002320 UP000038040 UP000274756 UP000008711 UP000095301 UP000069940 UP000249989 UP000095286 UP000037069 UP000008820 UP000007819 UP000009046
UP000030742 UP000019118 UP000235965 UP000027135 UP000002358 UP000215335 UP000007266 UP000053825 UP000008237 UP000079169 UP000053105 UP000092444 UP000078492 UP000078200 UP000007755 UP000075809 UP000076502 UP000092445 UP000078542 UP000005205 UP000078540 UP000242457 UP000005203 UP000078541 UP000092460 UP000092443 UP000053097 UP000279307 UP000008792 UP000000311 UP000008744 UP000001819 UP000268350 UP000001070 UP000095300 UP000009192 UP000015103 UP000002320 UP000038040 UP000274756 UP000008711 UP000095301 UP000069940 UP000249989 UP000095286 UP000037069 UP000008820 UP000007819 UP000009046
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9J6K0
A0A2H1V2N4
A0A2W1BFQ4
A0A194PGJ2
A0A2A4JW66
A0A194RKL8
+ More
A0A212F8Y0 A0A0L7LNU1 A0A2H1W4P3 U4UDM8 N6TR79 A0A2J7QCV6 V5H0D6 A0A067R639 K7J1I1 A0A2R7WF52 A0A232FIH3 A0A1Y1L0D7 D6WTA9 A0A0L7R1D6 E2C820 A0A3Q0ILT9 A0A0V0G6A5 E9IUM4 A0A0N0BDG4 A0A1B0GEU5 A0A195DUT2 A0A023F1Z6 A0A1A9VSM3 F4W8A6 A0A151WJA6 A0A154PIT6 A0A1A9ZM93 A0A195CCN0 A0A158NSN6 A0A151I6B5 A0A0C9R4X5 A0A2A3EQ80 A0A087ZZQ3 A0A0T6B8D9 A0A195EXK8 A0A1B0AVB9 A0A1A9YBN7 A0A026WXN3 A0A0Q9WPC2 A0A0Q9WQX2 A0A224X5V1 E2AKY7 B4LW20 A0A0P4VK91 A0A1B6CBG0 B4GP61 A0A0A9XUQ5 Q29BM2 A0A3B0K5A5 A0A146M558 B4JEK9 A0A1I8PWA3 A0A034VRR0 A0A023EW91 B4K4Z8 T1HHY4 B0WS34 A0A0N4UP28 A0A0K8V1K2 A0A1B6LSU4 B3NZS4 A0A1I8MUB4 A0A182GXK5 A0A0K8UCQ8 U5EZM2 A0A1B6IHW2 A0A2S2Q527 A0A0A1X4Y7 A0A1I8CJ16 A0A0L0C6C3 A0A1Q3FDG5 A0A1L8E1R2 Q16NG2 A0A1L8DR70 J9K9U5 A0A2S2P0E5 W8C710 W8BNH3 E0VV82 A0A2S2PN40 A0A1B6FR71
A0A212F8Y0 A0A0L7LNU1 A0A2H1W4P3 U4UDM8 N6TR79 A0A2J7QCV6 V5H0D6 A0A067R639 K7J1I1 A0A2R7WF52 A0A232FIH3 A0A1Y1L0D7 D6WTA9 A0A0L7R1D6 E2C820 A0A3Q0ILT9 A0A0V0G6A5 E9IUM4 A0A0N0BDG4 A0A1B0GEU5 A0A195DUT2 A0A023F1Z6 A0A1A9VSM3 F4W8A6 A0A151WJA6 A0A154PIT6 A0A1A9ZM93 A0A195CCN0 A0A158NSN6 A0A151I6B5 A0A0C9R4X5 A0A2A3EQ80 A0A087ZZQ3 A0A0T6B8D9 A0A195EXK8 A0A1B0AVB9 A0A1A9YBN7 A0A026WXN3 A0A0Q9WPC2 A0A0Q9WQX2 A0A224X5V1 E2AKY7 B4LW20 A0A0P4VK91 A0A1B6CBG0 B4GP61 A0A0A9XUQ5 Q29BM2 A0A3B0K5A5 A0A146M558 B4JEK9 A0A1I8PWA3 A0A034VRR0 A0A023EW91 B4K4Z8 T1HHY4 B0WS34 A0A0N4UP28 A0A0K8V1K2 A0A1B6LSU4 B3NZS4 A0A1I8MUB4 A0A182GXK5 A0A0K8UCQ8 U5EZM2 A0A1B6IHW2 A0A2S2Q527 A0A0A1X4Y7 A0A1I8CJ16 A0A0L0C6C3 A0A1Q3FDG5 A0A1L8E1R2 Q16NG2 A0A1L8DR70 J9K9U5 A0A2S2P0E5 W8C710 W8BNH3 E0VV82 A0A2S2PN40 A0A1B6FR71
PDB
1HXK
E-value=3.56768e-50,
Score=500
Ontologies
GO
Topology
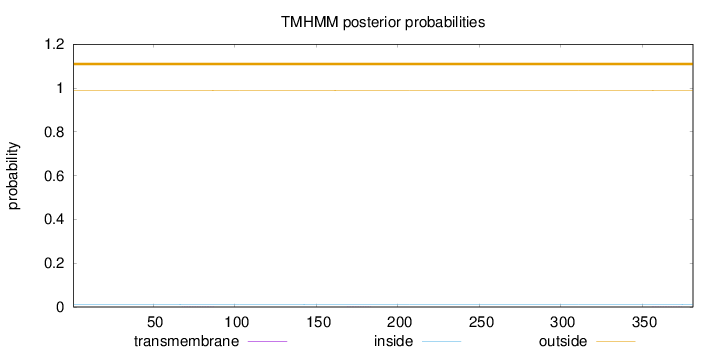
Length:
381
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01961
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.01092
outside
1 - 381
Population Genetic Test Statistics
Pi
202.042086
Theta
146.612386
Tajima's D
1.229923
CLR
0.021955
CSRT
0.71936403179841
Interpretation
Uncertain
