Gene
KWMTBOMO15310
Pre Gene Modal
BGIBMGA005057
Annotation
PREDICTED:_dihydroxyacetone_phosphate_acyltransferase_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.15 PlasmaMembrane Reliability : 1.623
Sequence
CDS
ATGAGTGAGGAAGTGAATTTCGTTGATATCCTAGAGCCGAGAAGGACTGAATCCGACACATTTCGCTACATGACTCGTAGGTGGGACCTTGTAAAAACTATGGCCTTAGATAAAATCTATTCGCCTGAAGATCTTAAAGATTTTGTGGCAAAATCGGTTTATTTAGACAATTTTATAGATGCTGAGAGCACCAGGACAGGTGTTAGCAAAACAGAATTGCACAAAGAAGTATTGAATTACCTTGAAGAAATGGGACTAGATAAAAAATTACATGTCATAAGATGGATGGGTGTCTTCTTCCTTAAAATTTCATTCATGATGAAGATCAAACTATTTGTCAATGAACCAAAAGTGCTCCAGCTAAAATTGACAATGGGAAGAAATCCTGTGTTGTTTCTTCCAACACATCGCAGTTATGCAGACTTTTGTCTTATGACCTATCTGTGTTGTCATTACGATATTGACTTTCCTGCTGTTGCTGCCGGAATGGATTTTTACTCAATGGCCATTGTGGGGCGACGTATGCGTGAAACGGGCGCTTTCTATATCAGGCGAACTTTAGTGGGTTCACCTCTGTATGCGGCAACATTGAGAGAGTATGTACGGACTGTAGTAGCCAAGTACTCATCTCCCATTGAATTCTTCCTAGAAGGCACAAGAAGCCGGAGCAATAAGAGCTTACCTCCAAAATACGGTATGCTTTCGATGAGCCTGATGCCGTACTTCGCTAGAGAAGTTAGCGACATTACCATAGTGCCCGTGAACATCAGCTACGACAGGCTCATGGAGCAGGGACTGTTCGCGTACGAACATCTCGGCGTGCCTAAACCCAAGGAATCCACTGGAGGTCTCTTGAAATCACTACGCAGCCTGAACGATCACTTCGGCAACATATACATCAACCTCGGCTCGCCGCTGTCCCTAAGAGGGTACTTGCAAGACGAATCGAGAGCCACCGAGGAAATAATGAAACCGAACGATCTCCAGCAAATTACGCCGGAACAATTCAGGCAGGTGCAGGACGTGGCGGAGCACGTGATAAGCCTGCAGCAACAGAGCACGGCGGTCACGATAACCAATCTAGTAGCTATAGTGCTCATGCAGAGCCTGGTGCGAGACGAGCCCCTGTCTCTCGATGGAGTCGTCGTTGAAGTGGTCTGGGTGGTCGGAGTGTTAGGAAAGCTGGGCGCTTCAGTGTTCGAGAATGACGTCAAAGGCAGCGTCGACAGGATCCTCGTGGTCCACAACAAGCTGCTGCGTGTCGATTCAAAACGAAAATTGAGACTGTTGTCGGAGACTTTGATGAACATGTCCAGTGAGGTCAAGAAAAAAATTAAAGGACACACTCTGGAAGCCGACACCATGGCCCTGGCCATACCAGTGATACAGCTGCAACTCTACGTCAACCCAGTAATGCACTATCTGTTTCCACCGGCTCTGGTGTACCTCATCGCCTCTAGAGGGGTAGACAGAGATATGTTGGTAACAGATTACAATCGCCTCAGGAAACTCTTTCGGAACGAATATTTTTACGTAGAAAAAAATGAAGAAAAGATTTTAAAAGAAGCAATAGAGTACTGCACGGCAAACGGCATCCTGATCTTCAACGGGGACAAATACAGTTGCGGGTCGGATGAGAAGTTGCAGAGGTTGTTGAAGTGGTCCGCGTGGCCGGCGCTCACCGTTGCCATTAAATGCGCTGAAATTATGACCGAGCAAACGACCTGCACACAAAAAGTAGCGCTGAAGCTGATACAGCAGCGGGTCGAGTCCCAGCGCTGCCACCCGTACTGCCTCAGTCTGGAGGCCGCGGCCGGCTGTCTGCGCGGGCTGCTCACGGTCGGAGCGCTGCACAGACAAAAGTGA
Protein
MSEEVNFVDILEPRRTESDTFRYMTRRWDLVKTMALDKIYSPEDLKDFVAKSVYLDNFIDAESTRTGVSKTELHKEVLNYLEEMGLDKKLHVIRWMGVFFLKISFMMKIKLFVNEPKVLQLKLTMGRNPVLFLPTHRSYADFCLMTYLCCHYDIDFPAVAAGMDFYSMAIVGRRMRETGAFYIRRTLVGSPLYAATLREYVRTVVAKYSSPIEFFLEGTRSRSNKSLPPKYGMLSMSLMPYFAREVSDITIVPVNISYDRLMEQGLFAYEHLGVPKPKESTGGLLKSLRSLNDHFGNIYINLGSPLSLRGYLQDESRATEEIMKPNDLQQITPEQFRQVQDVAEHVISLQQQSTAVTITNLVAIVLMQSLVRDEPLSLDGVVVEVVWVVGVLGKLGASVFENDVKGSVDRILVVHNKLLRVDSKRKLRLLSETLMNMSSEVKKKIKGHTLEADTMALAIPVIQLQLYVNPVMHYLFPPALVYLIASRGVDRDMLVTDYNRLRKLFRNEYFYVEKNEEKILKEAIEYCTANGILIFNGDKYSCGSDEKLQRLLKWSAWPALTVAIKCAEIMTEQTTCTQKVALKLIQQRVESQRCHPYCLSLEAAAGCLRGLLTVGALHRQK
Summary
Similarity
Belongs to the GPAT/DAPAT family.
Uniprot
H9J6B5
A0A2A4JUG1
A0A291P104
A0A291P198
A0A1V0M888
A0A194PGA6
+ More
A0A212F8X7 A0A088ML81 A0A1Y1NBK6 A0A084VRY4 D6WQ67 A0A067QQ32 A0A182JQR0 A0A1B6EVF1 A0A2J7Q4X3 A0A182N5Y6 A0A2J7Q4W7 A0A182MMH2 A0A2J7Q4X0 A0A023EUW6 A0A2J7Q4W5 A0A182PC34 A0A182GBH6 A0A1W4WXS3 Q16L82 A0A1Q3F3Q8 A0A182I0E4 A0A182YJT0 A0A182TUY7 A0A1Q3F3N8 A0A182V748 Q7PYF0 A0A182R1J8 A0A0L0BX70 A0A182VQZ0 A0A182XBK5 A0A2M4AI13 A0A182IWK3 A0A1B6CG76 A0A1B6BZ69 A0A1B0BUG8 A0A1Y9G8F3 A0A1Y9HB68 A0A1A9YCT1 A0A1A9Z3V3 A0A1A9V7X3 B0XKM6 A0A1Q3F3E6 E0W059 A0A1B6J208 A0A1I8PIM9 A0A2M4BFW1 A0A2M4BFU6 A0A1I8NB39 A0A1W4WM15 W5J691 A0A2M4CI57 B4NKS6 B4K4X3 A0A1A9WFM3 B4M4L8 A0A1B0FMH2 A0A336MYH8 A0A182KM33 A0A0T6B4N5 A0A336K3H7 B3LZ13 Q9VBQ6 B4PTK1 Q296H5 B4QUX0 B4ICA2 A0A1W4VM21 A0A3B0K653 A0A0B4KHK0 B3P6I8 B0X4B9 A0A0M4ET25 Q9NHF2 A0A3B0KBN9 A0A034W2G9 A0A0K8UN39 A0A0K8VTE6 W8AD89 A0A2P8YEH6 B4JHK6 A0A0A1XT50 B4GF53 A0A1I8PII9 A0A2T7NZ30 A0A0N7ZCG7 A0A195FQR9 A0A195BJ35
A0A212F8X7 A0A088ML81 A0A1Y1NBK6 A0A084VRY4 D6WQ67 A0A067QQ32 A0A182JQR0 A0A1B6EVF1 A0A2J7Q4X3 A0A182N5Y6 A0A2J7Q4W7 A0A182MMH2 A0A2J7Q4X0 A0A023EUW6 A0A2J7Q4W5 A0A182PC34 A0A182GBH6 A0A1W4WXS3 Q16L82 A0A1Q3F3Q8 A0A182I0E4 A0A182YJT0 A0A182TUY7 A0A1Q3F3N8 A0A182V748 Q7PYF0 A0A182R1J8 A0A0L0BX70 A0A182VQZ0 A0A182XBK5 A0A2M4AI13 A0A182IWK3 A0A1B6CG76 A0A1B6BZ69 A0A1B0BUG8 A0A1Y9G8F3 A0A1Y9HB68 A0A1A9YCT1 A0A1A9Z3V3 A0A1A9V7X3 B0XKM6 A0A1Q3F3E6 E0W059 A0A1B6J208 A0A1I8PIM9 A0A2M4BFW1 A0A2M4BFU6 A0A1I8NB39 A0A1W4WM15 W5J691 A0A2M4CI57 B4NKS6 B4K4X3 A0A1A9WFM3 B4M4L8 A0A1B0FMH2 A0A336MYH8 A0A182KM33 A0A0T6B4N5 A0A336K3H7 B3LZ13 Q9VBQ6 B4PTK1 Q296H5 B4QUX0 B4ICA2 A0A1W4VM21 A0A3B0K653 A0A0B4KHK0 B3P6I8 B0X4B9 A0A0M4ET25 Q9NHF2 A0A3B0KBN9 A0A034W2G9 A0A0K8UN39 A0A0K8VTE6 W8AD89 A0A2P8YEH6 B4JHK6 A0A0A1XT50 B4GF53 A0A1I8PII9 A0A2T7NZ30 A0A0N7ZCG7 A0A195FQR9 A0A195BJ35
Pubmed
19121390
28986331
28349297
26354079
22118469
26385554
+ More
28004739 24438588 18362917 19820115 24845553 24945155 26483478 17510324 25244985 12364791 26108605 20566863 25315136 20920257 23761445 17994087 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 25348373 24495485 29403074 25830018
28004739 24438588 18362917 19820115 24845553 24945155 26483478 17510324 25244985 12364791 26108605 20566863 25315136 20920257 23761445 17994087 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 25348373 24495485 29403074 25830018
EMBL
BABH01019831
BABH01019832
BABH01019833
BABH01019834
BABH01019835
BABH01019836
+ More
BABH01019837 NWSH01000562 PCG75641.1 MF687655 ATJ44581.1 MF706191 ATJ44618.1 KU755492 ARD71202.1 KQ459604 KPI92421.1 AGBW02009658 OWR50205.1 KJ579219 AIN34695.1 GEZM01009880 JAV94250.1 ATLV01015787 KE525036 KFB40728.1 KQ971354 EFA06119.2 KK853067 KDR11845.1 GECZ01028096 JAS41673.1 NEVH01018373 PNF23630.1 PNF23629.1 AXCM01002463 PNF23628.1 GAPW01000368 JAC13230.1 PNF23627.1 JXUM01158226 JXUM01158227 KQ572979 KXJ67969.1 CH477918 EAT35065.1 GFDL01012855 JAV22190.1 APCN01002072 GFDL01012871 JAV22174.1 AAAB01008987 EAA01039.5 JRES01001197 KNC24653.1 GGFK01007108 MBW40429.1 GEDC01027748 GEDC01024802 GEDC01022096 GEDC01011632 JAS09550.1 JAS12496.1 JAS15202.1 JAS25666.1 GEDC01030737 GEDC01030017 GEDC01025765 GEDC01022767 GEDC01021673 GEDC01015123 GEDC01006941 GEDC01005297 JAS06561.1 JAS07281.1 JAS11533.1 JAS14531.1 JAS15625.1 JAS22175.1 JAS30357.1 JAS32001.1 JXJN01020692 AXCN02000191 DS233904 EDS32179.1 GFDL01012965 JAV22080.1 DS235857 EEB19015.1 GECU01014485 JAS93221.1 GGFJ01002783 MBW51924.1 GGFJ01002784 MBW51925.1 ADMH02002134 ETN58375.1 GGFL01000846 MBW65024.1 CH964272 EDW84137.2 CH933806 EDW16126.2 CH940652 EDW59579.2 CCAG010018533 UFQT01003877 SSX35336.1 LJIG01009923 KRT82110.1 UFQS01000100 UFQT01000100 SSW99544.1 SSX19924.1 CH902617 EDV41887.2 AE014297 BT099931 AAF56473.2 ACX36507.1 AGB96336.1 CM000160 EDW98741.2 CM000070 EAL28482.3 CM000364 EDX14436.1 CH480828 EDW44998.1 OUUW01000007 SPP83550.1 AGB96337.1 CH954182 EDV53658.1 DS232337 EDS40272.1 CP012526 ALC47997.1 AF222991 AAF34698.1 SPP83549.1 GAKP01010977 GAKP01010974 GAKP01010971 GAKP01010968 JAC47978.1 GDHF01031688 GDHF01024308 JAI20626.1 JAI28006.1 GDHF01022361 GDHF01010504 JAI29953.1 JAI41810.1 GAMC01020589 GAMC01020587 GAMC01020585 GAMC01020584 JAB85970.1 PYGN01000661 PSN42636.1 CH916369 EDV92833.1 GBXI01000172 JAD14120.1 CH479182 EDW34238.1 PZQS01000008 PVD26429.1 GDRN01067210 JAI64400.1 KQ981305 KYN42930.1 KQ976455 KYM85111.1
BABH01019837 NWSH01000562 PCG75641.1 MF687655 ATJ44581.1 MF706191 ATJ44618.1 KU755492 ARD71202.1 KQ459604 KPI92421.1 AGBW02009658 OWR50205.1 KJ579219 AIN34695.1 GEZM01009880 JAV94250.1 ATLV01015787 KE525036 KFB40728.1 KQ971354 EFA06119.2 KK853067 KDR11845.1 GECZ01028096 JAS41673.1 NEVH01018373 PNF23630.1 PNF23629.1 AXCM01002463 PNF23628.1 GAPW01000368 JAC13230.1 PNF23627.1 JXUM01158226 JXUM01158227 KQ572979 KXJ67969.1 CH477918 EAT35065.1 GFDL01012855 JAV22190.1 APCN01002072 GFDL01012871 JAV22174.1 AAAB01008987 EAA01039.5 JRES01001197 KNC24653.1 GGFK01007108 MBW40429.1 GEDC01027748 GEDC01024802 GEDC01022096 GEDC01011632 JAS09550.1 JAS12496.1 JAS15202.1 JAS25666.1 GEDC01030737 GEDC01030017 GEDC01025765 GEDC01022767 GEDC01021673 GEDC01015123 GEDC01006941 GEDC01005297 JAS06561.1 JAS07281.1 JAS11533.1 JAS14531.1 JAS15625.1 JAS22175.1 JAS30357.1 JAS32001.1 JXJN01020692 AXCN02000191 DS233904 EDS32179.1 GFDL01012965 JAV22080.1 DS235857 EEB19015.1 GECU01014485 JAS93221.1 GGFJ01002783 MBW51924.1 GGFJ01002784 MBW51925.1 ADMH02002134 ETN58375.1 GGFL01000846 MBW65024.1 CH964272 EDW84137.2 CH933806 EDW16126.2 CH940652 EDW59579.2 CCAG010018533 UFQT01003877 SSX35336.1 LJIG01009923 KRT82110.1 UFQS01000100 UFQT01000100 SSW99544.1 SSX19924.1 CH902617 EDV41887.2 AE014297 BT099931 AAF56473.2 ACX36507.1 AGB96336.1 CM000160 EDW98741.2 CM000070 EAL28482.3 CM000364 EDX14436.1 CH480828 EDW44998.1 OUUW01000007 SPP83550.1 AGB96337.1 CH954182 EDV53658.1 DS232337 EDS40272.1 CP012526 ALC47997.1 AF222991 AAF34698.1 SPP83549.1 GAKP01010977 GAKP01010974 GAKP01010971 GAKP01010968 JAC47978.1 GDHF01031688 GDHF01024308 JAI20626.1 JAI28006.1 GDHF01022361 GDHF01010504 JAI29953.1 JAI41810.1 GAMC01020589 GAMC01020587 GAMC01020585 GAMC01020584 JAB85970.1 PYGN01000661 PSN42636.1 CH916369 EDV92833.1 GBXI01000172 JAD14120.1 CH479182 EDW34238.1 PZQS01000008 PVD26429.1 GDRN01067210 JAI64400.1 KQ981305 KYN42930.1 KQ976455 KYM85111.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000030765
UP000007266
+ More
UP000027135 UP000075881 UP000235965 UP000075884 UP000075883 UP000075885 UP000069940 UP000249989 UP000192223 UP000008820 UP000075840 UP000076408 UP000075902 UP000075903 UP000007062 UP000075900 UP000037069 UP000075920 UP000076407 UP000075880 UP000092460 UP000069272 UP000075886 UP000092443 UP000092445 UP000078200 UP000002320 UP000009046 UP000095300 UP000095301 UP000000673 UP000007798 UP000009192 UP000091820 UP000008792 UP000092444 UP000075882 UP000007801 UP000000803 UP000002282 UP000001819 UP000000304 UP000001292 UP000192221 UP000268350 UP000008711 UP000092553 UP000245037 UP000001070 UP000008744 UP000245119 UP000078541 UP000078540
UP000027135 UP000075881 UP000235965 UP000075884 UP000075883 UP000075885 UP000069940 UP000249989 UP000192223 UP000008820 UP000075840 UP000076408 UP000075902 UP000075903 UP000007062 UP000075900 UP000037069 UP000075920 UP000076407 UP000075880 UP000092460 UP000069272 UP000075886 UP000092443 UP000092445 UP000078200 UP000002320 UP000009046 UP000095300 UP000095301 UP000000673 UP000007798 UP000009192 UP000091820 UP000008792 UP000092444 UP000075882 UP000007801 UP000000803 UP000002282 UP000001819 UP000000304 UP000001292 UP000192221 UP000268350 UP000008711 UP000092553 UP000245037 UP000001070 UP000008744 UP000245119 UP000078541 UP000078540
Interpro
Gene 3D
ProteinModelPortal
H9J6B5
A0A2A4JUG1
A0A291P104
A0A291P198
A0A1V0M888
A0A194PGA6
+ More
A0A212F8X7 A0A088ML81 A0A1Y1NBK6 A0A084VRY4 D6WQ67 A0A067QQ32 A0A182JQR0 A0A1B6EVF1 A0A2J7Q4X3 A0A182N5Y6 A0A2J7Q4W7 A0A182MMH2 A0A2J7Q4X0 A0A023EUW6 A0A2J7Q4W5 A0A182PC34 A0A182GBH6 A0A1W4WXS3 Q16L82 A0A1Q3F3Q8 A0A182I0E4 A0A182YJT0 A0A182TUY7 A0A1Q3F3N8 A0A182V748 Q7PYF0 A0A182R1J8 A0A0L0BX70 A0A182VQZ0 A0A182XBK5 A0A2M4AI13 A0A182IWK3 A0A1B6CG76 A0A1B6BZ69 A0A1B0BUG8 A0A1Y9G8F3 A0A1Y9HB68 A0A1A9YCT1 A0A1A9Z3V3 A0A1A9V7X3 B0XKM6 A0A1Q3F3E6 E0W059 A0A1B6J208 A0A1I8PIM9 A0A2M4BFW1 A0A2M4BFU6 A0A1I8NB39 A0A1W4WM15 W5J691 A0A2M4CI57 B4NKS6 B4K4X3 A0A1A9WFM3 B4M4L8 A0A1B0FMH2 A0A336MYH8 A0A182KM33 A0A0T6B4N5 A0A336K3H7 B3LZ13 Q9VBQ6 B4PTK1 Q296H5 B4QUX0 B4ICA2 A0A1W4VM21 A0A3B0K653 A0A0B4KHK0 B3P6I8 B0X4B9 A0A0M4ET25 Q9NHF2 A0A3B0KBN9 A0A034W2G9 A0A0K8UN39 A0A0K8VTE6 W8AD89 A0A2P8YEH6 B4JHK6 A0A0A1XT50 B4GF53 A0A1I8PII9 A0A2T7NZ30 A0A0N7ZCG7 A0A195FQR9 A0A195BJ35
A0A212F8X7 A0A088ML81 A0A1Y1NBK6 A0A084VRY4 D6WQ67 A0A067QQ32 A0A182JQR0 A0A1B6EVF1 A0A2J7Q4X3 A0A182N5Y6 A0A2J7Q4W7 A0A182MMH2 A0A2J7Q4X0 A0A023EUW6 A0A2J7Q4W5 A0A182PC34 A0A182GBH6 A0A1W4WXS3 Q16L82 A0A1Q3F3Q8 A0A182I0E4 A0A182YJT0 A0A182TUY7 A0A1Q3F3N8 A0A182V748 Q7PYF0 A0A182R1J8 A0A0L0BX70 A0A182VQZ0 A0A182XBK5 A0A2M4AI13 A0A182IWK3 A0A1B6CG76 A0A1B6BZ69 A0A1B0BUG8 A0A1Y9G8F3 A0A1Y9HB68 A0A1A9YCT1 A0A1A9Z3V3 A0A1A9V7X3 B0XKM6 A0A1Q3F3E6 E0W059 A0A1B6J208 A0A1I8PIM9 A0A2M4BFW1 A0A2M4BFU6 A0A1I8NB39 A0A1W4WM15 W5J691 A0A2M4CI57 B4NKS6 B4K4X3 A0A1A9WFM3 B4M4L8 A0A1B0FMH2 A0A336MYH8 A0A182KM33 A0A0T6B4N5 A0A336K3H7 B3LZ13 Q9VBQ6 B4PTK1 Q296H5 B4QUX0 B4ICA2 A0A1W4VM21 A0A3B0K653 A0A0B4KHK0 B3P6I8 B0X4B9 A0A0M4ET25 Q9NHF2 A0A3B0KBN9 A0A034W2G9 A0A0K8UN39 A0A0K8VTE6 W8AD89 A0A2P8YEH6 B4JHK6 A0A0A1XT50 B4GF53 A0A1I8PII9 A0A2T7NZ30 A0A0N7ZCG7 A0A195FQR9 A0A195BJ35
Ontologies
PATHWAY
GO
PANTHER
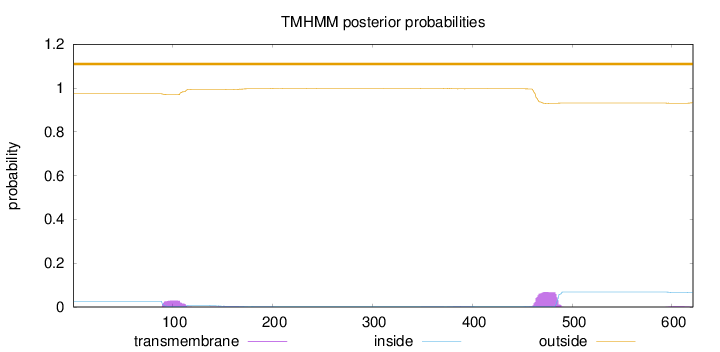
Topology
Length:
621
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.25946
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02627
outside
1 - 621
Population Genetic Test Statistics
Pi
2.383653
Theta
18.626323
Tajima's D
1.0664
CLR
0.526924
CSRT
0.688365581720914
Interpretation
Uncertain
