Gene
KWMTBOMO15309
Pre Gene Modal
BGIBMGA005055
Annotation
PREDICTED:_putative_helicase_Mov10l1_isoform_X1_[Bombyx_mori]
Full name
Putative helicase mov-10-B.1
+ More
Putative helicase mov-10-B.2
Putative helicase mov-10-B.2
Location in the cell
Nuclear Reliability : 1.658 PlasmaMembrane Reliability : 1.121
Sequence
CDS
ATGAATTGCCAGTTGTGTTCTGAAAGCAATGTACCTTCTGATCATCACACTACTGAGAGACATATGAAAAATAAGACTTTGTTTGATTACTCTGTATACAAAAAACAGAATGTTGGAAATAAGAAGGGTATCATTGTATCATGTGATGTTAGATCTTCAAGCTCCACACATTCTCAGTGTTATGAGTCAAAAATAAATGTTTCTGTTAAGCCAAATGAACAAGTAGAGTTCACTTTCACAATCTTAAATGACACCAAGAAGGACAAATTTTCAATCAATTGTGCTCAAATAATGCATTCCAAATTAAACTTTACTATCAATGATCACGGAATTGATGCAGGCTCTCCACCACACATCATCAAGCCCAAATCGAAGCTCAACAAGAAAATTACAGTGATGTTCAGATCAGCGAGCGTTGGAGAATATGTGATGCCAATAGTATTCTGTTTTATTATGAACAGTACAAATGAAAACATTGTTATCGTTAGGGACATGGTCGTTTACGTTGAAAAACAGAAAAAGCTTCACGACAAAATGTACAGTCCTTTTGTACTTCAAAGTATTGTGGCAGAAAAGACATACAGATGCGTCCGTGAGGGGACAAGAGAAGAGGGTATATTCACAGTGCCAAAGAATTACAAGCCTATTTACAGTTCCGCGTTAAAGTGCCCCTCTAATCTCACTCAAGAAAAAGCAAAGCTGGTGGCAGATTTAAGGAAAATTTTCAGTACTGGCATCACCAAAATGAACTATGTTCAGTTTTTTCATCATCTGCTCTGGTACGAAGAAACTGTCCTCAAGAATAATCTAAAGAATTACAACATGACAGGCGTGAATCTAAAGCTCGGTAGCATTCCATCGTCGTACACGTTGGTCGTGCCCGGCTTAGCGGAGAAAAGACCGTCGTTGTTGGTTGGTGATTATTTGTTCGTTAAACCGCAAAATGAGAAGATAATGTTCGAAGCGAACGTCACTCTAATAACCGAGAATGACGTCACTATACAAGGCATGCATTCAACCTTTTTTAAGAACTACTCTAAAACTAAGAAGTACGACATACGGTTCACGATGTCCCGCGTGACCTTGGAACGGATGCACCAGGCGGTACACTTCGCCGAACAACAGATGCCCAGATTGTTCCCTACGCACTATACAAATAAGAAGAAAGTAGAACCAATAACGGAATTTTTTAACGTGAACATCCGAGACAACTTGGAGCAGCGCTCAGCCGTGGAGCACATCGTGTCGGGCAGCAGCGGGCGCGCGCCCTACCTCGTGCACGGGCCGCCCGGCACCGGCAAGACCGTCACCATCGTCGAAGCCATCCTGCAGCTTGTGGTGCGGAACTCTAAAAATCGAATACTTGTCTGCACCGACTCCAACATGGCGGCGGATAACGTGGCCACGATGCTTATAAAGTACGCCAAAAAGTTTCCTTCCGAAAAGTTTCTTTTAAGAGCGAGCTCCTTTTATCGAACATGGGCAACACTACCAGAATGCTTACATGCCTTTTCGAACGGTACAAGTACCAAAGACTTTTGGTCTGTGAGTTTCACAGTTTTCCAGAAATACGCGATCGTCATTACAACGTTATCGCACGCCGCCAAATTCAGCGGAAGTTTTAGTAAGAATAGCACGTGCCACATAACGCATCTGTTCATCGACGAAGCGGCCCAGGCTTCCGAGCCTGCCAGTCTCATCCCGATCTGCGGGCTGTTGTCTACCGCTGGTTCCCTGGTACTGGCCGGGGATCCCCTGCAACTGGGACCTGTCATCATTTCACAAAAGGCCAAAGAAATTGGCCTCGGTCTATCGCTCATGGAACGTTTGATGAAAACGTTTCCTCAGTACAACGACGTGAAGAACGACTGCAACTATATAATGCTGCTGCGCAATAACTTCAGATCTGATCCGGACATACTGCACATCCCTAACGAGTTGTTCTACAAGGGACAGCTCAGAGCTCATGCTAGTGAAGATCCGATAAGCAAGACCGATTTATCCGGCGAGAAACCATCCAGTCGCGCTATTATGTTCCACGGGGTCCTGTCTAAGGAACAGAAAGTTGGCATGTCACCGAGCTTCTACAATAACTCTGAGTTAGAAATCGTCCAGATGTACATAACGTTTCTCGTGAACGATCATAGAATCCAACTGGAAGATATCGGCGTCATCACTCCGTACATCCGACAAGCCTATCACATCAAGAAATGGCTCTCGAGCAATAATTACGAAAAAATTGAAGTCGGCACAGTCGAGTCGTTCCAAGGGAAGGAAAAGCGCGTCATCGTCGTGTCCACGGTTCGAGCCAACAACGACCTCCTCGCGCACGACGCCAAGTTCCAGCTCGGCTTCCTGGTTGATGATAAGCGTTTCAATGTGGCGCTGACACGTGCGAAAGCCAAACTTATCATTATTGGCAACCCCTTGTGTCTCCGGAAGGACAAGAAGTGGCAACTCTACATGCAGATGTGCCGCGAACTTGGCACCTATTTTGGTTTTGACAGTGACAACATTGAAACCGACGAACTGGACCGAAAGGTCGATGAGATCCTGCCGCTTTTCAGACACATTGAGGTTTCAAAACAAGAATAG
Protein
MNCQLCSESNVPSDHHTTERHMKNKTLFDYSVYKKQNVGNKKGIIVSCDVRSSSSTHSQCYESKINVSVKPNEQVEFTFTILNDTKKDKFSINCAQIMHSKLNFTINDHGIDAGSPPHIIKPKSKLNKKITVMFRSASVGEYVMPIVFCFIMNSTNENIVIVRDMVVYVEKQKKLHDKMYSPFVLQSIVAEKTYRCVREGTREEGIFTVPKNYKPIYSSALKCPSNLTQEKAKLVADLRKIFSTGITKMNYVQFFHHLLWYEETVLKNNLKNYNMTGVNLKLGSIPSSYTLVVPGLAEKRPSLLVGDYLFVKPQNEKIMFEANVTLITENDVTIQGMHSTFFKNYSKTKKYDIRFTMSRVTLERMHQAVHFAEQQMPRLFPTHYTNKKKVEPITEFFNVNIRDNLEQRSAVEHIVSGSSGRAPYLVHGPPGTGKTVTIVEAILQLVVRNSKNRILVCTDSNMAADNVATMLIKYAKKFPSEKFLLRASSFYRTWATLPECLHAFSNGTSTKDFWSVSFTVFQKYAIVITTLSHAAKFSGSFSKNSTCHITHLFIDEAAQASEPASLIPICGLLSTAGSLVLAGDPLQLGPVIISQKAKEIGLGLSLMERLMKTFPQYNDVKNDCNYIMLLRNNFRSDPDILHIPNELFYKGQLRAHASEDPISKTDLSGEKPSSRAIMFHGVLSKEQKVGMSPSFYNNSELEIVQMYITFLVNDHRIQLEDIGVITPYIRQAYHIKKWLSSNNYEKIEVGTVESFQGKEKRVIVVSTVRANNDLLAHDAKFQLGFLVDDKRFNVALTRAKAKLIIIGNPLCLRKDKKWQLYMQMCRELGTYFGFDSDNIETDELDRKVDEILPLFRHIEVSKQE
Summary
Description
Probable RNA helicase. Required for RNA-mediated gene silencing by the RNA-induced silencing complex (RISC). Required for both miRNA-mediated translational repression and miRNA-mediated cleavage of complementary mRNAs by RISC (By similarity).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the DNA2/NAM7 helicase family. SDE3 subfamily.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Helicase
Hydrolase
Nucleotide-binding
Reference proteome
RNA-binding
RNA-mediated gene silencing
Feature
chain Putative helicase mov-10-B.1
Uniprot
A0A2H1VG04
A0A2A4JW82
A0A2W1BH77
A0A194PMS1
A0A194RL89
A0A3S2NN90
+ More
A0A212F8Y4 A0A1Y1LYA7 A0A2J7RK32 A0A1Y1LUS6 D6WC75 A0A2P8XCY3 V5GVH9 V5GJG8 A0A067RGC4 A0A1S3JER6 C3YAE9 Q1LXK4 A0A3P9P4N1 A0A3B3YTV1 A0A3B3XJF3 E7FAV9 A0A3Q2QVI1 A0A210QWW2 T1IYZ0 A0A146WTF8 A0A3B5PS47 A0A3P9P524 A0A3B3Z2B2 A0A146NPF4 A0A147BNX8 A0A131Y1W2 A0A3Q0S277 A0A3B3V1Y8 A0A3B3UZQ6 A0A2R8QPU7 A0A3B3UZP7 A0A3B3Q6V6 A0A3B3VLH2 A0A2R5LCG3 A0A3B4DPV0 A0A2G8LF59 A0A3B3ZT81 A0A067QX16 A0A087X566 A0A2I4B8M6 A0A371D0W8 A0A1W4ZBZ1 A0A3Q2U397 I3K2W3 M3ZF91 A0A146P8M9 A0A3B3C6G2 A0A1A8PTL8 A0A1Z5L1L7 A0A3P9Q782 A0A0C5XIN6 A0A3P9DQ48 A0A0P7UK81 A0A3P9KYN0 A0A1A8V552 I3K2W4 A0A1A8BHP7 A0A3P8RR32 A0A3B4GH31 A0A3Q4I2S6 A0A3P8VNH1 A0A3P8P1Y6 A0A3P9ILM6 A0A2G8JWJ6 A0A096M4W4 A0A1A8LBT4 A0A1A8NEX2 A0A3Q3ATU1 H2VC76 G1KUK0 K1QA54 A0A1A7YE69 A0A2I4C0Y2 A0A1W2W4F1 A0A3Q3LVL9 A0A1A7WDM5 A0A3B5BEL0 A0A3P9KB63 A0A1A8UTU0 A0A1A8EIU2 A0A1A8FFE9 A0A3Q2FDY9 W5MW54 T1IMR9 A0A3Q1G8A4 A0A3M6UH79 Q1LXK5 A0A3Q2DM66
A0A212F8Y4 A0A1Y1LYA7 A0A2J7RK32 A0A1Y1LUS6 D6WC75 A0A2P8XCY3 V5GVH9 V5GJG8 A0A067RGC4 A0A1S3JER6 C3YAE9 Q1LXK4 A0A3P9P4N1 A0A3B3YTV1 A0A3B3XJF3 E7FAV9 A0A3Q2QVI1 A0A210QWW2 T1IYZ0 A0A146WTF8 A0A3B5PS47 A0A3P9P524 A0A3B3Z2B2 A0A146NPF4 A0A147BNX8 A0A131Y1W2 A0A3Q0S277 A0A3B3V1Y8 A0A3B3UZQ6 A0A2R8QPU7 A0A3B3UZP7 A0A3B3Q6V6 A0A3B3VLH2 A0A2R5LCG3 A0A3B4DPV0 A0A2G8LF59 A0A3B3ZT81 A0A067QX16 A0A087X566 A0A2I4B8M6 A0A371D0W8 A0A1W4ZBZ1 A0A3Q2U397 I3K2W3 M3ZF91 A0A146P8M9 A0A3B3C6G2 A0A1A8PTL8 A0A1Z5L1L7 A0A3P9Q782 A0A0C5XIN6 A0A3P9DQ48 A0A0P7UK81 A0A3P9KYN0 A0A1A8V552 I3K2W4 A0A1A8BHP7 A0A3P8RR32 A0A3B4GH31 A0A3Q4I2S6 A0A3P8VNH1 A0A3P8P1Y6 A0A3P9ILM6 A0A2G8JWJ6 A0A096M4W4 A0A1A8LBT4 A0A1A8NEX2 A0A3Q3ATU1 H2VC76 G1KUK0 K1QA54 A0A1A7YE69 A0A2I4C0Y2 A0A1W2W4F1 A0A3Q3LVL9 A0A1A7WDM5 A0A3B5BEL0 A0A3P9KB63 A0A1A8UTU0 A0A1A8EIU2 A0A1A8FFE9 A0A3Q2FDY9 W5MW54 T1IMR9 A0A3Q1G8A4 A0A3M6UH79 Q1LXK5 A0A3Q2DM66
EC Number
3.6.4.13
Pubmed
EMBL
ODYU01002367
SOQ39778.1
NWSH01000562
PCG75642.1
KZ150095
PZC73621.1
+ More
KQ459604 KPI92420.1 KQ460045 KPJ18085.1 RSAL01000001 RVE55283.1 AGBW02009658 OWR50204.1 GEZM01046296 JAV77350.1 NEVH01002981 PNF41192.1 GEZM01046297 JAV77349.1 KQ971316 EEZ97839.2 PYGN01002902 PSN29852.1 GALX01004228 JAB64238.1 GALX01004227 JAB64239.1 KK852657 KDR19239.1 GG666494 EEN62697.1 BX323059 BC171375 CU469362 NEDP02001463 OWF53196.1 JH431701 GCES01053125 JAR33198.1 GCES01152608 JAQ33714.1 GEGO01002951 JAR92453.1 GEFM01002313 JAP73483.1 BX248319 GGLE01003019 MBY07145.1 MRZV01000101 PIK58790.1 KK852866 KDR14736.1 AYCK01012747 KZ857430 RDX46173.1 AERX01019455 AERX01019456 AERX01019457 GCES01146592 JAQ39730.1 HAEI01003374 SBR84329.1 GFJQ02005687 JAW01283.1 KP128064 AJR21491.1 JARO02009229 KPP61866.1 HADY01017972 HAEJ01015033 SBS55490.1 HADZ01002235 SBP66176.1 MRZV01001162 PIK40113.1 HAEF01004576 SBR41958.1 HAEH01001676 SBR67377.1 JH818778 EKC30838.1 HADX01006207 SBP28439.1 HADW01002513 SBP03913.1 HADY01012439 HAEJ01011033 SBS51490.1 HAEB01000573 SBQ47025.1 HAEB01011225 SBQ57752.1 AHAT01014313 JH431094 RCHS01001556 RMX52929.1
KQ459604 KPI92420.1 KQ460045 KPJ18085.1 RSAL01000001 RVE55283.1 AGBW02009658 OWR50204.1 GEZM01046296 JAV77350.1 NEVH01002981 PNF41192.1 GEZM01046297 JAV77349.1 KQ971316 EEZ97839.2 PYGN01002902 PSN29852.1 GALX01004228 JAB64238.1 GALX01004227 JAB64239.1 KK852657 KDR19239.1 GG666494 EEN62697.1 BX323059 BC171375 CU469362 NEDP02001463 OWF53196.1 JH431701 GCES01053125 JAR33198.1 GCES01152608 JAQ33714.1 GEGO01002951 JAR92453.1 GEFM01002313 JAP73483.1 BX248319 GGLE01003019 MBY07145.1 MRZV01000101 PIK58790.1 KK852866 KDR14736.1 AYCK01012747 KZ857430 RDX46173.1 AERX01019455 AERX01019456 AERX01019457 GCES01146592 JAQ39730.1 HAEI01003374 SBR84329.1 GFJQ02005687 JAW01283.1 KP128064 AJR21491.1 JARO02009229 KPP61866.1 HADY01017972 HAEJ01015033 SBS55490.1 HADZ01002235 SBP66176.1 MRZV01001162 PIK40113.1 HAEF01004576 SBR41958.1 HAEH01001676 SBR67377.1 JH818778 EKC30838.1 HADX01006207 SBP28439.1 HADW01002513 SBP03913.1 HADY01012439 HAEJ01011033 SBS51490.1 HAEB01000573 SBQ47025.1 HAEB01011225 SBQ57752.1 AHAT01014313 JH431094 RCHS01001556 RMX52929.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
UP000235965
+ More
UP000007266 UP000245037 UP000027135 UP000085678 UP000001554 UP000000437 UP000242638 UP000261480 UP000265000 UP000242188 UP000002852 UP000261340 UP000261500 UP000261540 UP000261440 UP000230750 UP000261520 UP000028760 UP000192220 UP000256964 UP000192224 UP000005207 UP000261560 UP000265160 UP000034805 UP000265180 UP000265080 UP000261460 UP000261580 UP000265120 UP000265100 UP000265200 UP000264800 UP000005226 UP000001646 UP000005408 UP000261660 UP000261400 UP000265020 UP000018468 UP000257200 UP000275408
UP000007266 UP000245037 UP000027135 UP000085678 UP000001554 UP000000437 UP000242638 UP000261480 UP000265000 UP000242188 UP000002852 UP000261340 UP000261500 UP000261540 UP000261440 UP000230750 UP000261520 UP000028760 UP000192220 UP000256964 UP000192224 UP000005207 UP000261560 UP000265160 UP000034805 UP000265180 UP000265080 UP000261460 UP000261580 UP000265120 UP000265100 UP000265200 UP000264800 UP000005226 UP000001646 UP000005408 UP000261660 UP000261400 UP000265020 UP000018468 UP000257200 UP000275408
Interpro
ProteinModelPortal
A0A2H1VG04
A0A2A4JW82
A0A2W1BH77
A0A194PMS1
A0A194RL89
A0A3S2NN90
+ More
A0A212F8Y4 A0A1Y1LYA7 A0A2J7RK32 A0A1Y1LUS6 D6WC75 A0A2P8XCY3 V5GVH9 V5GJG8 A0A067RGC4 A0A1S3JER6 C3YAE9 Q1LXK4 A0A3P9P4N1 A0A3B3YTV1 A0A3B3XJF3 E7FAV9 A0A3Q2QVI1 A0A210QWW2 T1IYZ0 A0A146WTF8 A0A3B5PS47 A0A3P9P524 A0A3B3Z2B2 A0A146NPF4 A0A147BNX8 A0A131Y1W2 A0A3Q0S277 A0A3B3V1Y8 A0A3B3UZQ6 A0A2R8QPU7 A0A3B3UZP7 A0A3B3Q6V6 A0A3B3VLH2 A0A2R5LCG3 A0A3B4DPV0 A0A2G8LF59 A0A3B3ZT81 A0A067QX16 A0A087X566 A0A2I4B8M6 A0A371D0W8 A0A1W4ZBZ1 A0A3Q2U397 I3K2W3 M3ZF91 A0A146P8M9 A0A3B3C6G2 A0A1A8PTL8 A0A1Z5L1L7 A0A3P9Q782 A0A0C5XIN6 A0A3P9DQ48 A0A0P7UK81 A0A3P9KYN0 A0A1A8V552 I3K2W4 A0A1A8BHP7 A0A3P8RR32 A0A3B4GH31 A0A3Q4I2S6 A0A3P8VNH1 A0A3P8P1Y6 A0A3P9ILM6 A0A2G8JWJ6 A0A096M4W4 A0A1A8LBT4 A0A1A8NEX2 A0A3Q3ATU1 H2VC76 G1KUK0 K1QA54 A0A1A7YE69 A0A2I4C0Y2 A0A1W2W4F1 A0A3Q3LVL9 A0A1A7WDM5 A0A3B5BEL0 A0A3P9KB63 A0A1A8UTU0 A0A1A8EIU2 A0A1A8FFE9 A0A3Q2FDY9 W5MW54 T1IMR9 A0A3Q1G8A4 A0A3M6UH79 Q1LXK5 A0A3Q2DM66
A0A212F8Y4 A0A1Y1LYA7 A0A2J7RK32 A0A1Y1LUS6 D6WC75 A0A2P8XCY3 V5GVH9 V5GJG8 A0A067RGC4 A0A1S3JER6 C3YAE9 Q1LXK4 A0A3P9P4N1 A0A3B3YTV1 A0A3B3XJF3 E7FAV9 A0A3Q2QVI1 A0A210QWW2 T1IYZ0 A0A146WTF8 A0A3B5PS47 A0A3P9P524 A0A3B3Z2B2 A0A146NPF4 A0A147BNX8 A0A131Y1W2 A0A3Q0S277 A0A3B3V1Y8 A0A3B3UZQ6 A0A2R8QPU7 A0A3B3UZP7 A0A3B3Q6V6 A0A3B3VLH2 A0A2R5LCG3 A0A3B4DPV0 A0A2G8LF59 A0A3B3ZT81 A0A067QX16 A0A087X566 A0A2I4B8M6 A0A371D0W8 A0A1W4ZBZ1 A0A3Q2U397 I3K2W3 M3ZF91 A0A146P8M9 A0A3B3C6G2 A0A1A8PTL8 A0A1Z5L1L7 A0A3P9Q782 A0A0C5XIN6 A0A3P9DQ48 A0A0P7UK81 A0A3P9KYN0 A0A1A8V552 I3K2W4 A0A1A8BHP7 A0A3P8RR32 A0A3B4GH31 A0A3Q4I2S6 A0A3P8VNH1 A0A3P8P1Y6 A0A3P9ILM6 A0A2G8JWJ6 A0A096M4W4 A0A1A8LBT4 A0A1A8NEX2 A0A3Q3ATU1 H2VC76 G1KUK0 K1QA54 A0A1A7YE69 A0A2I4C0Y2 A0A1W2W4F1 A0A3Q3LVL9 A0A1A7WDM5 A0A3B5BEL0 A0A3P9KB63 A0A1A8UTU0 A0A1A8EIU2 A0A1A8FFE9 A0A3Q2FDY9 W5MW54 T1IMR9 A0A3Q1G8A4 A0A3M6UH79 Q1LXK5 A0A3Q2DM66
PDB
2WJY
E-value=1.00296e-36,
Score=388
Ontologies
GO
PANTHER
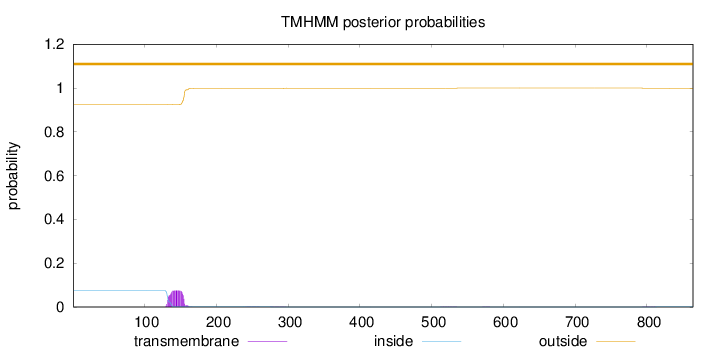
Topology
Subcellular location
Length:
864
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.67425
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07530
outside
1 - 864
Population Genetic Test Statistics
Pi
338.013445
Theta
190.189247
Tajima's D
1.288007
CLR
0.619495
CSRT
0.738613069346533
Interpretation
Uncertain
