Gene
KWMTBOMO15307
Pre Gene Modal
BGIBMGA005143
Annotation
PREDICTED:_uncharacterized_protein_LOC106721700_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.946
Sequence
CDS
ATGGGGATGATTTCAAGAGTGCGCGGGCGCATTAGAAGTGATTCGTTCACAAAAATTCACACACTCAAATCAGAGGACAAAATTGCGAATATTGTATGGCTTCTTTCACTTGTTTTCCTGTTGCCATTTTGGGCTCCACGAGGATTACTGGTTGCACTTGGAGGCGCATGGCTGATGGCAGCGTACACATGTTTAGTTGTTCCTGTACTAATATCAGCGAACTACCGGTATCCAGCCAGATGGTATCCAGCGGTAGCGAAAATAGTCCGAAGAGTAGCCAGAAAGCTGCGCCCATATTTTGACTGTGCCAAGAATGTTTTAAGGGTGGCGTATGCACCACTAGAACGTACAGAGAAATTATTTTTACAGATGAACAATCTCTCAACTATGATGTGTCAGTTGCTGATGTTCACAGCTTGTGATCGACTATTTCTCTCACAAGCAAAACTAACCAGTCTGTATTCCTTGATGTTCTATAACGTAGTCACCTATTCAGTTAGCTACATTCGTGAGATCTGCACGAAGGAAGACTGGTCGCCGTTTGTAAACGTAACACGACATTCCCAAGTAAAGCACCTCGCGATGTCTGCGACTAAAATCGTCCTCGAATGGACGAAAGCCATCACATTCATAGTGACAATCACTTTTATACTCCTTGCCTTTGGATTAGAACAAGGCTTGGAGCACTATCGGCCGACGGCACTGTACACCGTGACTACCGGCATCTATTACTTGTTATCAGAGAGAACGTTTTTAGAATTGTGGCCGATTGCTATTTCGGCGCTAAAGCTAGAAAGACTAGAAGGCATGGAAACATTGTACTTCGGAGTTTGGGCGCGGGGTGTGACTACAGCGTTGGCCGTGCCTCTAGTGCCAGCTTTGGCCTGGTGTGGCCGTTGGCGATTGGCTATATTGGTGCTGTACCTGTGTGTATTCGTTCACGGCAGACATCGTCTCGGAGAAGCGCTGATAAAGTTGAATGAAGCTAGAGGGTCCCTGGCGAAATTCAGACGTGCAACTTCACAGGAGCTGGCCACTTTGGAAGACGTTTGCGCCGTGTGCCTAGGTGCCATGAGGTCTGCACGAGTCACGCCTTGCGCTCACTATTTCCACGCTGACTGCTTGAGAAGATGCCTTGCCGCTTCGGATAGATGCCCCATTTGTGTCAGACCTTATGTATTTTGCTGA
Protein
MGMISRVRGRIRSDSFTKIHTLKSEDKIANIVWLLSLVFLLPFWAPRGLLVALGGAWLMAAYTCLVVPVLISANYRYPARWYPAVAKIVRRVARKLRPYFDCAKNVLRVAYAPLERTEKLFLQMNNLSTMMCQLLMFTACDRLFLSQAKLTSLYSLMFYNVVTYSVSYIREICTKEDWSPFVNVTRHSQVKHLAMSATKIVLEWTKAITFIVTITFILLAFGLEQGLEHYRPTALYTVTTGIYYLLSERTFLELWPIAISALKLERLEGMETLYFGVWARGVTTALAVPLVPALAWCGRWRLAILVLYLCVFVHGRHRLGEALIKLNEARGSLAKFRRATSQELATLEDVCAVCLGAMRSARVTPCAHYFHADCLRRCLAASDRCPICVRPYVFC
Summary
Uniprot
A0A194RKM3
A0A3S2LWK1
A0A194PI66
A0A2W1BPL0
A0A2H1VG60
A0A212F8Y6
+ More
A0A0L7LIY3 A0A139WH61 A0A067R401 A0A1B6K443 A0A1B6JAG1 A0A1Y1LUH8 A0A1W4WYG9 A0A2P8XML8 A0A0C9Q6W5 A0A1B6MLX8 A0A195C8B2 E1ZXL5 A0A195BGT8 A0A158P047 E9ISY6 A0A026WPG8 F4X3T8 A0A195F282 A0A195DZX3 A0A151XJK7 E2BLK0 A0A0L7QXS3 A0A0J7L1H1 U4UMW3 N6U0D1 A0A2R7W8F8 A0A232ERB7 A0A0A9XQN7 A0A088A2Z1 T1INZ9 A0A1B6F204 T1HHK5 K7JI91 A0A1B6DMJ4 A0A1B6C7I7 A0A293MXG3 A0A1Z5KWU9 A0A2R5LA60 A0A3Q0J538 A0A1E1XJZ1 A0A1E1XGQ9 A0A226DXE8 L7MD84 A0A1D2MVI3 A0A1W4WYM0 N6TRW2 U4UG75 D6WME6 A0A067QLC0 A0A2P8YBG7 A0A2J7QH93 A0A2J7QHA0 A0A1B6LLC7 A0A1B6DQU8
A0A0L7LIY3 A0A139WH61 A0A067R401 A0A1B6K443 A0A1B6JAG1 A0A1Y1LUH8 A0A1W4WYG9 A0A2P8XML8 A0A0C9Q6W5 A0A1B6MLX8 A0A195C8B2 E1ZXL5 A0A195BGT8 A0A158P047 E9ISY6 A0A026WPG8 F4X3T8 A0A195F282 A0A195DZX3 A0A151XJK7 E2BLK0 A0A0L7QXS3 A0A0J7L1H1 U4UMW3 N6U0D1 A0A2R7W8F8 A0A232ERB7 A0A0A9XQN7 A0A088A2Z1 T1INZ9 A0A1B6F204 T1HHK5 K7JI91 A0A1B6DMJ4 A0A1B6C7I7 A0A293MXG3 A0A1Z5KWU9 A0A2R5LA60 A0A3Q0J538 A0A1E1XJZ1 A0A1E1XGQ9 A0A226DXE8 L7MD84 A0A1D2MVI3 A0A1W4WYM0 N6TRW2 U4UG75 D6WME6 A0A067QLC0 A0A2P8YBG7 A0A2J7QH93 A0A2J7QHA0 A0A1B6LLC7 A0A1B6DQU8
Pubmed
EMBL
KQ460045
KPJ18087.1
RSAL01000001
RVE55281.1
KQ459604
KPI92419.1
+ More
KZ150095 PZC73623.1 ODYU01002367 SOQ39776.1 AGBW02009658 OWR50202.1 JTDY01000926 KOB75407.1 KQ971343 KYB27302.1 KK852718 KDR17789.1 GECU01008508 GECU01001477 JAS99198.1 JAT06230.1 GECU01011485 JAS96221.1 GEZM01047960 JAV76681.1 PYGN01001711 PSN33247.1 GBYB01009848 JAG79615.1 GEBQ01008308 GEBQ01003098 JAT31669.1 JAT36879.1 KQ978081 KYM97072.1 GL435066 EFN74098.1 KQ976490 KYM83367.1 ADTU01005352 ADTU01005353 GL765434 EFZ16299.1 KK107139 QOIP01000003 EZA57868.1 RLU24668.1 GL888624 EGI58885.1 KQ981856 KYN34491.1 KQ979955 KYN18460.1 KQ982074 KYQ60517.1 GL449033 EFN83437.1 KQ414704 KOC63351.1 LBMM01001273 KMQ96607.1 KB632281 ERL91335.1 APGK01054087 KB741247 ENN71997.1 KK854419 PTY15708.1 NNAY01002666 OXU20826.1 GBHO01044024 GBHO01022489 GBHO01022487 GBHO01022486 GBHO01022485 GBRD01015079 GBRD01015077 GBRD01015076 GBRD01015075 GBRD01015074 GBRD01015073 GBRD01015072 GBRD01001090 GDHC01020012 GDHC01015492 GDHC01007018 JAF99579.1 JAG21115.1 JAG21117.1 JAG21118.1 JAG21119.1 JAG50747.1 JAP98616.1 JAQ03137.1 JAQ11611.1 JH431232 GECZ01025565 JAS44204.1 ACPB03011203 GEDC01010420 JAS26878.1 GEDC01028073 JAS09225.1 GFWV01020784 MAA45512.1 GFJQ02007532 JAV99437.1 GGLE01002209 MBY06335.1 GFAA01004008 JAT99426.1 GFAC01000967 JAT98221.1 LNIX01000009 OXA49943.1 GACK01002853 JAA62181.1 LJIJ01000475 ODM97069.1 APGK01054088 ENN72000.1 ERL91338.1 EFA04260.2 KK853186 KDR10081.1 PYGN01000730 PSN41586.1 NEVH01013984 PNF27954.1 PNF27956.1 GEBQ01015432 JAT24545.1 GEDC01009234 JAS28064.1
KZ150095 PZC73623.1 ODYU01002367 SOQ39776.1 AGBW02009658 OWR50202.1 JTDY01000926 KOB75407.1 KQ971343 KYB27302.1 KK852718 KDR17789.1 GECU01008508 GECU01001477 JAS99198.1 JAT06230.1 GECU01011485 JAS96221.1 GEZM01047960 JAV76681.1 PYGN01001711 PSN33247.1 GBYB01009848 JAG79615.1 GEBQ01008308 GEBQ01003098 JAT31669.1 JAT36879.1 KQ978081 KYM97072.1 GL435066 EFN74098.1 KQ976490 KYM83367.1 ADTU01005352 ADTU01005353 GL765434 EFZ16299.1 KK107139 QOIP01000003 EZA57868.1 RLU24668.1 GL888624 EGI58885.1 KQ981856 KYN34491.1 KQ979955 KYN18460.1 KQ982074 KYQ60517.1 GL449033 EFN83437.1 KQ414704 KOC63351.1 LBMM01001273 KMQ96607.1 KB632281 ERL91335.1 APGK01054087 KB741247 ENN71997.1 KK854419 PTY15708.1 NNAY01002666 OXU20826.1 GBHO01044024 GBHO01022489 GBHO01022487 GBHO01022486 GBHO01022485 GBRD01015079 GBRD01015077 GBRD01015076 GBRD01015075 GBRD01015074 GBRD01015073 GBRD01015072 GBRD01001090 GDHC01020012 GDHC01015492 GDHC01007018 JAF99579.1 JAG21115.1 JAG21117.1 JAG21118.1 JAG21119.1 JAG50747.1 JAP98616.1 JAQ03137.1 JAQ11611.1 JH431232 GECZ01025565 JAS44204.1 ACPB03011203 GEDC01010420 JAS26878.1 GEDC01028073 JAS09225.1 GFWV01020784 MAA45512.1 GFJQ02007532 JAV99437.1 GGLE01002209 MBY06335.1 GFAA01004008 JAT99426.1 GFAC01000967 JAT98221.1 LNIX01000009 OXA49943.1 GACK01002853 JAA62181.1 LJIJ01000475 ODM97069.1 APGK01054088 ENN72000.1 ERL91338.1 EFA04260.2 KK853186 KDR10081.1 PYGN01000730 PSN41586.1 NEVH01013984 PNF27954.1 PNF27956.1 GEBQ01015432 JAT24545.1 GEDC01009234 JAS28064.1
Proteomes
UP000053240
UP000283053
UP000053268
UP000007151
UP000037510
UP000007266
+ More
UP000027135 UP000192223 UP000245037 UP000078542 UP000000311 UP000078540 UP000005205 UP000053097 UP000279307 UP000007755 UP000078541 UP000078492 UP000075809 UP000008237 UP000053825 UP000036403 UP000030742 UP000019118 UP000215335 UP000005203 UP000015103 UP000002358 UP000079169 UP000198287 UP000094527 UP000235965
UP000027135 UP000192223 UP000245037 UP000078542 UP000000311 UP000078540 UP000005205 UP000053097 UP000279307 UP000007755 UP000078541 UP000078492 UP000075809 UP000008237 UP000053825 UP000036403 UP000030742 UP000019118 UP000215335 UP000005203 UP000015103 UP000002358 UP000079169 UP000198287 UP000094527 UP000235965
Interpro
SUPFAM
SSF46689
SSF46689
Gene 3D
ProteinModelPortal
A0A194RKM3
A0A3S2LWK1
A0A194PI66
A0A2W1BPL0
A0A2H1VG60
A0A212F8Y6
+ More
A0A0L7LIY3 A0A139WH61 A0A067R401 A0A1B6K443 A0A1B6JAG1 A0A1Y1LUH8 A0A1W4WYG9 A0A2P8XML8 A0A0C9Q6W5 A0A1B6MLX8 A0A195C8B2 E1ZXL5 A0A195BGT8 A0A158P047 E9ISY6 A0A026WPG8 F4X3T8 A0A195F282 A0A195DZX3 A0A151XJK7 E2BLK0 A0A0L7QXS3 A0A0J7L1H1 U4UMW3 N6U0D1 A0A2R7W8F8 A0A232ERB7 A0A0A9XQN7 A0A088A2Z1 T1INZ9 A0A1B6F204 T1HHK5 K7JI91 A0A1B6DMJ4 A0A1B6C7I7 A0A293MXG3 A0A1Z5KWU9 A0A2R5LA60 A0A3Q0J538 A0A1E1XJZ1 A0A1E1XGQ9 A0A226DXE8 L7MD84 A0A1D2MVI3 A0A1W4WYM0 N6TRW2 U4UG75 D6WME6 A0A067QLC0 A0A2P8YBG7 A0A2J7QH93 A0A2J7QHA0 A0A1B6LLC7 A0A1B6DQU8
A0A0L7LIY3 A0A139WH61 A0A067R401 A0A1B6K443 A0A1B6JAG1 A0A1Y1LUH8 A0A1W4WYG9 A0A2P8XML8 A0A0C9Q6W5 A0A1B6MLX8 A0A195C8B2 E1ZXL5 A0A195BGT8 A0A158P047 E9ISY6 A0A026WPG8 F4X3T8 A0A195F282 A0A195DZX3 A0A151XJK7 E2BLK0 A0A0L7QXS3 A0A0J7L1H1 U4UMW3 N6U0D1 A0A2R7W8F8 A0A232ERB7 A0A0A9XQN7 A0A088A2Z1 T1INZ9 A0A1B6F204 T1HHK5 K7JI91 A0A1B6DMJ4 A0A1B6C7I7 A0A293MXG3 A0A1Z5KWU9 A0A2R5LA60 A0A3Q0J538 A0A1E1XJZ1 A0A1E1XGQ9 A0A226DXE8 L7MD84 A0A1D2MVI3 A0A1W4WYM0 N6TRW2 U4UG75 D6WME6 A0A067QLC0 A0A2P8YBG7 A0A2J7QH93 A0A2J7QHA0 A0A1B6LLC7 A0A1B6DQU8
PDB
4LAD
E-value=6.36927e-09,
Score=145
Ontologies
GO
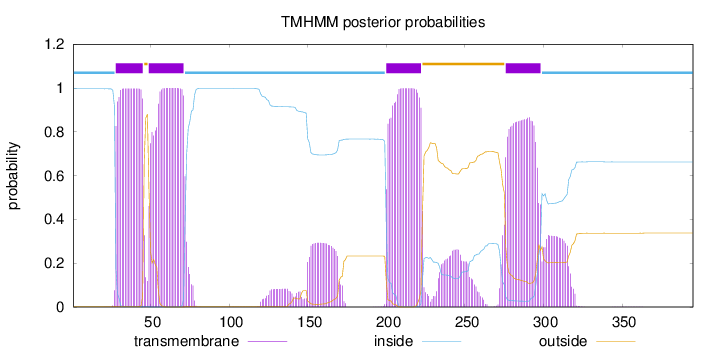
Topology
Length:
395
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
101.52745
Exp number, first 60 AAs:
28.60422
Total prob of N-in:
0.99670
POSSIBLE N-term signal
sequence
inside
1 - 27
TMhelix
28 - 45
outside
46 - 48
TMhelix
49 - 71
inside
72 - 199
TMhelix
200 - 222
outside
223 - 275
TMhelix
276 - 298
inside
299 - 395
Population Genetic Test Statistics
Pi
25.722454
Theta
223.453579
Tajima's D
0.30789
CLR
0.528441
CSRT
0.456527173641318
Interpretation
Uncertain
