Gene
KWMTBOMO15301
Pre Gene Modal
BGIBMGA005147
Annotation
PREDICTED:_anaphase-promoting_complex_subunit_2_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.67 Nuclear Reliability : 1.812
Sequence
CDS
ATGGGCGAGGACATAATGTGTTCTGGTTGTGGAAACGAGTATTCCGATTGTAACTGTGCATACATTGTGAAAGTATTTCATGAAACAAATAGGAAGTTGGTGGAGTTACAATTGTTAGAAAGACTCACAGGTCAAGTTCTAACTAATTTCATCCAGTTGAGAATTCAGTCTCATATACAGAAAGTATGTACTGGCACTTTTGATGTGTCCCACATTGAAGCATTAGAAAGTTGGCTTAATACGACAGTAATGTCTTGGTTGATCAGAATTTATTGTGCTGGCTCCTCCAAACCTCCACCTGATGATAAAAATATACAAAATGCAATCTCAAGATTCAAACAGAAACTCACTTACTATCTCTATCATAGCTACACAAAAACAAGAATTGATCAGCTCTTCAACATCATTATAGAATATCCTGATTCCCAGCCTGCTGTTGATGACATCAAAATTTGTTTAGCAAAAACAGATTTGAGACCAACTCTATGTAAAAAGTTACAGAATGCATTAGAAACAAGGCTTCTTCACCCAGGCGTGAACACTCCTGACATACTCACTGCTTACATATCTACTATTAGAGCTTTGAGACACCTAGATCCTTCTGGTGTTATACTGGAAACAGTGACAAAACCTGTTCGTAATTATTTAAGAAGTAGAGAAGATACAGTGAGGAGTGTTGTGAGCAGTTTGACAGAAGAAGGTGCTGGTAGTGAGTTAGCTGAAGAGCTAGCTAAATTTGCAGCAGAAGGAGATGGCGATGCCGATAAAGATGAGGCTTGGGATGAATGGAATCCTGATCCAGTTGATGCTGATCCCAAAGTTGATTCAAATGACAGAAGAACTAATGATATTATATCTATGCTTGTTAATGTTTATGGTAGCAAGGAATTGTTTGTTAATGAATATAGAACTTTACTAGCTGATAGATTACTGGCTCAAAGTGTTATCAATACAGAAAAAGAAATTAGGTACCTGGAGCTATTAAAAATAAGATTTGGTGAATCACAATTACACTTCTGTGAAGTCATGCTTAAAGATATATCAGATTCAAAGAGAATTAATTCCTTAATACATCAGGACAAAACCATTGAGGAGCTGAATAAGAAATTTACTTCTAATGCAATGATTTTATCTGCACAGTTTTGGCCACCATTTAAAGATGAAAGTCTGGAATTACCAGAAGAAATCAAGGAACATTTTGAAGCATACACTAAATCTTATGAAGCTTTAAAAGGTAATCGAACATTAAGCTGGAAACCACACCTTGGAAATGTAAACTTAGAGATAGAAATAGGCGAAAAGAAGTTAGATCTGACAGTTTCACCTTTTAATGCGACATTGATAATGCATTTCCAAAATAAATCTGAATGGACGTTAGATGAACTCCATCAGGTGATGAAAGTTCCTATTACTATTCTGAGAAGGAAAATAACATACTGGCAATCGATGGGTTTAATAGCAGAAAAAAGTACAGATCACTTTGTTTTAGTTGAAGCTGATAATAAAGCCAGTGTACCTTCACATCAAGTACAGGAAATGATTTGTGAAGATGAAGAAACGGAGAGTGCCATGGCATCAGCACATGATCAGCGGGAAGGTGAACTGCAAGTTTTTTGGTCTTATATTGTAGGCATGTTAACAAACTTGGACTGTTTGCCTTTGGACAGAATACATCAAATGTTGAAGATGTTTGCTTCCCAAGCTCCAGGCACTGAATGCAGTCTTATGGAGCTGAGACAATTTTTAGACACAAAAGTAAGAACACATCAACTGGTACTTCAAGGAGGCATGTACAAATTGCCTAAGACATAG
Protein
MGEDIMCSGCGNEYSDCNCAYIVKVFHETNRKLVELQLLERLTGQVLTNFIQLRIQSHIQKVCTGTFDVSHIEALESWLNTTVMSWLIRIYCAGSSKPPPDDKNIQNAISRFKQKLTYYLYHSYTKTRIDQLFNIIIEYPDSQPAVDDIKICLAKTDLRPTLCKKLQNALETRLLHPGVNTPDILTAYISTIRALRHLDPSGVILETVTKPVRNYLRSREDTVRSVVSSLTEEGAGSELAEELAKFAAEGDGDADKDEAWDEWNPDPVDADPKVDSNDRRTNDIISMLVNVYGSKELFVNEYRTLLADRLLAQSVINTEKEIRYLELLKIRFGESQLHFCEVMLKDISDSKRINSLIHQDKTIEELNKKFTSNAMILSAQFWPPFKDESLELPEEIKEHFEAYTKSYEALKGNRTLSWKPHLGNVNLEIEIGEKKLDLTVSPFNATLIMHFQNKSEWTLDELHQVMKVPITILRRKITYWQSMGLIAEKSTDHFVLVEADNKASVPSHQVQEMICEDEETESAMASAHDQREGELQVFWSYIVGMLTNLDCLPLDRIHQMLKMFASQAPGTECSLMELRQFLDTKVRTHQLVLQGGMYKLPKT
Summary
Similarity
Belongs to the cullin family.
Uniprot
A0A2W1BDL6
A0A2H1VNG1
A0A2A4JCQ6
A0A0L7LM05
A0A212F8W4
A0A194RL01
+ More
H9J6K5 A0A2J7PN50 A0A067R8L3 A0A1B6GKW8 A0A1B6FIH1 A0A1B6GF83 A0A194PMR1 E2ACD3 A0A023F112 A0A0A9Y699 T1HNQ8 A0A232EZD5 K7J566 A0A0J7L0T9 F4WZV1 A0A195FED4 A0A158NGM3 A0A151I8P4 A0A195B7Z7 A0A2A3EPT4 A0A0P4VWJ7 A0A088ADT4 A0A195DXK2 A0A151XAG8 A0A026WZF5 A0A0C9R8F9 E9IA33 A0A0L7QJW7 A0A1W4XK67 E0VT63 E2C9F0 A0A1B6HAS7 D6WHQ9 A0A1Y1LLF6 A0A154PHW4 A0A1W4XL28 A0A210Q1Q3 K1Q363 A0A1S3KB87 A0A1S3HPQ2 A0A1W4UZQ9 A0A1J1IRR5 Q9W1E5 B4QBF9 Q290C0 B3NQ41 B4GCG5 A0A3B0IZ35 B3MCP3 A0A3B0JIX6 A0A3B0IYV1 B4PAB8 B4J680 B4I8Y3 A0A336M6D5 A0A336KH44 A0A146L3R1 A0A3R7MSA7 A0A0L8I8M4 A0A1I8NG42 B4MRL5 B4KLY2 A0A1I8P1Z1 B4LP09 V4CRB5 A0A2H8TG57 A0A0M4EIL6 A0A0L0BQL8 T1IXP7 A0A0P4VYU1 A0A0P4W136 A0A0A1X7P6 A0A1B0CUP5 A0A0K8U519 A0A0K8UK47 A0A087TDE9 W8C8Q2 A0A2C9KE64 V9KFN7 W4YRS9 A0A2L2YGV9 A0A1A9ZZH8 A0A1A9V697 A0A1B0BW34 A0A2B4T1Z3 A0A0K2TV75 A0A1A9X9L8 A0A1A9X5T1 A0A293LQV3 A0A0T6BCR5 A0A099YR40 A0A2R5LDU8 M7ATY9 A0A091NHU4
H9J6K5 A0A2J7PN50 A0A067R8L3 A0A1B6GKW8 A0A1B6FIH1 A0A1B6GF83 A0A194PMR1 E2ACD3 A0A023F112 A0A0A9Y699 T1HNQ8 A0A232EZD5 K7J566 A0A0J7L0T9 F4WZV1 A0A195FED4 A0A158NGM3 A0A151I8P4 A0A195B7Z7 A0A2A3EPT4 A0A0P4VWJ7 A0A088ADT4 A0A195DXK2 A0A151XAG8 A0A026WZF5 A0A0C9R8F9 E9IA33 A0A0L7QJW7 A0A1W4XK67 E0VT63 E2C9F0 A0A1B6HAS7 D6WHQ9 A0A1Y1LLF6 A0A154PHW4 A0A1W4XL28 A0A210Q1Q3 K1Q363 A0A1S3KB87 A0A1S3HPQ2 A0A1W4UZQ9 A0A1J1IRR5 Q9W1E5 B4QBF9 Q290C0 B3NQ41 B4GCG5 A0A3B0IZ35 B3MCP3 A0A3B0JIX6 A0A3B0IYV1 B4PAB8 B4J680 B4I8Y3 A0A336M6D5 A0A336KH44 A0A146L3R1 A0A3R7MSA7 A0A0L8I8M4 A0A1I8NG42 B4MRL5 B4KLY2 A0A1I8P1Z1 B4LP09 V4CRB5 A0A2H8TG57 A0A0M4EIL6 A0A0L0BQL8 T1IXP7 A0A0P4VYU1 A0A0P4W136 A0A0A1X7P6 A0A1B0CUP5 A0A0K8U519 A0A0K8UK47 A0A087TDE9 W8C8Q2 A0A2C9KE64 V9KFN7 W4YRS9 A0A2L2YGV9 A0A1A9ZZH8 A0A1A9V697 A0A1B0BW34 A0A2B4T1Z3 A0A0K2TV75 A0A1A9X9L8 A0A1A9X5T1 A0A293LQV3 A0A0T6BCR5 A0A099YR40 A0A2R5LDU8 M7ATY9 A0A091NHU4
Pubmed
28756777
26227816
22118469
26354079
19121390
24845553
+ More
20798317 25474469 25401762 28648823 20075255 21719571 21347285 27129103 24508170 30249741 21282665 20566863 18362917 19820115 28004739 28812685 22992520 10731132 12537568 12537572 12537573 12537574 12169670 16110336 17569856 17569867 26109357 26109356 17994087 22936249 15632085 17550304 26823975 25315136 23254933 26108605 25830018 24495485 15562597 24402279 26561354 23624526
20798317 25474469 25401762 28648823 20075255 21719571 21347285 27129103 24508170 30249741 21282665 20566863 18362917 19820115 28004739 28812685 22992520 10731132 12537568 12537572 12537573 12537574 12169670 16110336 17569856 17569867 26109357 26109356 17994087 22936249 15632085 17550304 26823975 25315136 23254933 26108605 25830018 24495485 15562597 24402279 26561354 23624526
EMBL
KZ150398
PZC71026.1
ODYU01003492
SOQ42360.1
NWSH01001968
PCG69536.1
+ More
JTDY01000685 KOB76226.1 AGBW02009658 OWR50195.1 KQ460045 KPJ18094.1 BABH01019861 BABH01019862 NEVH01023957 PNF17762.1 KK852811 KDR15929.1 GECZ01006738 JAS63031.1 GECZ01019825 JAS49944.1 GECZ01008654 JAS61115.1 KQ459604 KPI92410.1 GL438491 EFN68923.1 GBBI01003510 JAC15202.1 GBHO01040684 GBHO01040683 GBHO01040681 GBHO01040680 GBHO01040679 GBHO01040671 GBHO01016454 GBHO01016448 JAG02920.1 JAG02921.1 JAG02923.1 JAG02924.1 JAG02925.1 JAG02933.1 JAG27150.1 JAG27156.1 ACPB03010549 NNAY01001588 OXU23508.1 AAZX01005033 LBMM01001479 KMQ96246.1 GL888480 EGI60241.1 KQ981625 KYN39040.1 ADTU01015145 KQ978346 KYM94765.1 KQ976574 KYM80319.1 KZ288206 PBC33051.1 GDKW01001522 JAI55073.1 KQ980107 KYN17608.1 KQ982339 KYQ57362.1 KK107063 QOIP01000005 EZA61203.1 RLU23081.1 GBYB01004340 JAG74107.1 GL761961 EFZ22570.1 KQ415041 KOC58879.1 DS235759 EEB16569.1 GL453837 EFN75428.1 GECU01035901 JAS71805.1 KQ971321 EFA00073.1 GEZM01055087 JAV73160.1 KQ434916 KZC11445.1 NEDP02005240 OWF42674.1 JH817484 EKC30892.1 CVRI01000058 CRL02911.1 AE013599 AY129297 BT031326 AAF47123.1 AAM97765.1 ABY21739.1 CM000362 CM002911 EDX08477.1 KMY96196.1 CM000071 EAL25442.1 CH954179 EDV56914.1 CH479181 EDW32442.1 OUUW01000001 SPP73197.1 CH902619 EDV36277.1 SPP73196.1 SPP73195.1 CM000158 EDW92444.1 CH916367 EDW01938.1 CH480824 EDW57058.1 UFQT01000521 SSX24921.1 UFQS01000465 UFQT01000465 SSX04236.1 SSX24601.1 GDHC01017219 GDHC01007839 JAQ01410.1 JAQ10790.1 QCYY01000517 ROT84629.1 KQ416263 KOF97749.1 CH963850 EDW74754.2 CH933808 EDW09792.1 CH940648 EDW61178.1 KB199651 ESP05035.1 GFXV01001292 MBW13097.1 CP012524 ALC42647.1 JRES01001518 KNC22283.1 JH431659 GDRN01097233 JAI59164.1 GDRN01097234 JAI59163.1 GBXI01007376 JAD06916.1 AJWK01029484 AJWK01029485 AJWK01029486 GDHF01030648 JAI21666.1 GDHF01025255 JAI27059.1 KK114715 KFM63138.1 GAMC01002834 JAC03722.1 JW864170 AFO96687.1 AAGJ04157311 IAAA01029192 IAAA01029193 LAA07384.1 JXJN01021585 LSMT01000002 PFX34802.1 HACA01012231 CDW29592.1 GFWV01005468 MAA30198.1 LJIG01002245 KRT84695.1 KL885225 KGL72774.1 GGLE01003530 MBY07656.1 KB579997 EMP26420.1 KK847204 KFP88582.1
JTDY01000685 KOB76226.1 AGBW02009658 OWR50195.1 KQ460045 KPJ18094.1 BABH01019861 BABH01019862 NEVH01023957 PNF17762.1 KK852811 KDR15929.1 GECZ01006738 JAS63031.1 GECZ01019825 JAS49944.1 GECZ01008654 JAS61115.1 KQ459604 KPI92410.1 GL438491 EFN68923.1 GBBI01003510 JAC15202.1 GBHO01040684 GBHO01040683 GBHO01040681 GBHO01040680 GBHO01040679 GBHO01040671 GBHO01016454 GBHO01016448 JAG02920.1 JAG02921.1 JAG02923.1 JAG02924.1 JAG02925.1 JAG02933.1 JAG27150.1 JAG27156.1 ACPB03010549 NNAY01001588 OXU23508.1 AAZX01005033 LBMM01001479 KMQ96246.1 GL888480 EGI60241.1 KQ981625 KYN39040.1 ADTU01015145 KQ978346 KYM94765.1 KQ976574 KYM80319.1 KZ288206 PBC33051.1 GDKW01001522 JAI55073.1 KQ980107 KYN17608.1 KQ982339 KYQ57362.1 KK107063 QOIP01000005 EZA61203.1 RLU23081.1 GBYB01004340 JAG74107.1 GL761961 EFZ22570.1 KQ415041 KOC58879.1 DS235759 EEB16569.1 GL453837 EFN75428.1 GECU01035901 JAS71805.1 KQ971321 EFA00073.1 GEZM01055087 JAV73160.1 KQ434916 KZC11445.1 NEDP02005240 OWF42674.1 JH817484 EKC30892.1 CVRI01000058 CRL02911.1 AE013599 AY129297 BT031326 AAF47123.1 AAM97765.1 ABY21739.1 CM000362 CM002911 EDX08477.1 KMY96196.1 CM000071 EAL25442.1 CH954179 EDV56914.1 CH479181 EDW32442.1 OUUW01000001 SPP73197.1 CH902619 EDV36277.1 SPP73196.1 SPP73195.1 CM000158 EDW92444.1 CH916367 EDW01938.1 CH480824 EDW57058.1 UFQT01000521 SSX24921.1 UFQS01000465 UFQT01000465 SSX04236.1 SSX24601.1 GDHC01017219 GDHC01007839 JAQ01410.1 JAQ10790.1 QCYY01000517 ROT84629.1 KQ416263 KOF97749.1 CH963850 EDW74754.2 CH933808 EDW09792.1 CH940648 EDW61178.1 KB199651 ESP05035.1 GFXV01001292 MBW13097.1 CP012524 ALC42647.1 JRES01001518 KNC22283.1 JH431659 GDRN01097233 JAI59164.1 GDRN01097234 JAI59163.1 GBXI01007376 JAD06916.1 AJWK01029484 AJWK01029485 AJWK01029486 GDHF01030648 JAI21666.1 GDHF01025255 JAI27059.1 KK114715 KFM63138.1 GAMC01002834 JAC03722.1 JW864170 AFO96687.1 AAGJ04157311 IAAA01029192 IAAA01029193 LAA07384.1 JXJN01021585 LSMT01000002 PFX34802.1 HACA01012231 CDW29592.1 GFWV01005468 MAA30198.1 LJIG01002245 KRT84695.1 KL885225 KGL72774.1 GGLE01003530 MBY07656.1 KB579997 EMP26420.1 KK847204 KFP88582.1
Proteomes
UP000218220
UP000037510
UP000007151
UP000053240
UP000005204
UP000235965
+ More
UP000027135 UP000053268 UP000000311 UP000015103 UP000215335 UP000002358 UP000036403 UP000007755 UP000078541 UP000005205 UP000078542 UP000078540 UP000242457 UP000005203 UP000078492 UP000075809 UP000053097 UP000279307 UP000053825 UP000192223 UP000009046 UP000008237 UP000007266 UP000076502 UP000242188 UP000005408 UP000085678 UP000192221 UP000183832 UP000000803 UP000000304 UP000001819 UP000008711 UP000008744 UP000268350 UP000007801 UP000002282 UP000001070 UP000001292 UP000283509 UP000053454 UP000095301 UP000007798 UP000009192 UP000095300 UP000008792 UP000030746 UP000092553 UP000037069 UP000092461 UP000054359 UP000076420 UP000007110 UP000092445 UP000078200 UP000092460 UP000225706 UP000092443 UP000091820 UP000053641 UP000031443
UP000027135 UP000053268 UP000000311 UP000015103 UP000215335 UP000002358 UP000036403 UP000007755 UP000078541 UP000005205 UP000078542 UP000078540 UP000242457 UP000005203 UP000078492 UP000075809 UP000053097 UP000279307 UP000053825 UP000192223 UP000009046 UP000008237 UP000007266 UP000076502 UP000242188 UP000005408 UP000085678 UP000192221 UP000183832 UP000000803 UP000000304 UP000001819 UP000008711 UP000008744 UP000268350 UP000007801 UP000002282 UP000001070 UP000001292 UP000283509 UP000053454 UP000095301 UP000007798 UP000009192 UP000095300 UP000008792 UP000030746 UP000092553 UP000037069 UP000092461 UP000054359 UP000076420 UP000007110 UP000092445 UP000078200 UP000092460 UP000225706 UP000092443 UP000091820 UP000053641 UP000031443
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BDL6
A0A2H1VNG1
A0A2A4JCQ6
A0A0L7LM05
A0A212F8W4
A0A194RL01
+ More
H9J6K5 A0A2J7PN50 A0A067R8L3 A0A1B6GKW8 A0A1B6FIH1 A0A1B6GF83 A0A194PMR1 E2ACD3 A0A023F112 A0A0A9Y699 T1HNQ8 A0A232EZD5 K7J566 A0A0J7L0T9 F4WZV1 A0A195FED4 A0A158NGM3 A0A151I8P4 A0A195B7Z7 A0A2A3EPT4 A0A0P4VWJ7 A0A088ADT4 A0A195DXK2 A0A151XAG8 A0A026WZF5 A0A0C9R8F9 E9IA33 A0A0L7QJW7 A0A1W4XK67 E0VT63 E2C9F0 A0A1B6HAS7 D6WHQ9 A0A1Y1LLF6 A0A154PHW4 A0A1W4XL28 A0A210Q1Q3 K1Q363 A0A1S3KB87 A0A1S3HPQ2 A0A1W4UZQ9 A0A1J1IRR5 Q9W1E5 B4QBF9 Q290C0 B3NQ41 B4GCG5 A0A3B0IZ35 B3MCP3 A0A3B0JIX6 A0A3B0IYV1 B4PAB8 B4J680 B4I8Y3 A0A336M6D5 A0A336KH44 A0A146L3R1 A0A3R7MSA7 A0A0L8I8M4 A0A1I8NG42 B4MRL5 B4KLY2 A0A1I8P1Z1 B4LP09 V4CRB5 A0A2H8TG57 A0A0M4EIL6 A0A0L0BQL8 T1IXP7 A0A0P4VYU1 A0A0P4W136 A0A0A1X7P6 A0A1B0CUP5 A0A0K8U519 A0A0K8UK47 A0A087TDE9 W8C8Q2 A0A2C9KE64 V9KFN7 W4YRS9 A0A2L2YGV9 A0A1A9ZZH8 A0A1A9V697 A0A1B0BW34 A0A2B4T1Z3 A0A0K2TV75 A0A1A9X9L8 A0A1A9X5T1 A0A293LQV3 A0A0T6BCR5 A0A099YR40 A0A2R5LDU8 M7ATY9 A0A091NHU4
H9J6K5 A0A2J7PN50 A0A067R8L3 A0A1B6GKW8 A0A1B6FIH1 A0A1B6GF83 A0A194PMR1 E2ACD3 A0A023F112 A0A0A9Y699 T1HNQ8 A0A232EZD5 K7J566 A0A0J7L0T9 F4WZV1 A0A195FED4 A0A158NGM3 A0A151I8P4 A0A195B7Z7 A0A2A3EPT4 A0A0P4VWJ7 A0A088ADT4 A0A195DXK2 A0A151XAG8 A0A026WZF5 A0A0C9R8F9 E9IA33 A0A0L7QJW7 A0A1W4XK67 E0VT63 E2C9F0 A0A1B6HAS7 D6WHQ9 A0A1Y1LLF6 A0A154PHW4 A0A1W4XL28 A0A210Q1Q3 K1Q363 A0A1S3KB87 A0A1S3HPQ2 A0A1W4UZQ9 A0A1J1IRR5 Q9W1E5 B4QBF9 Q290C0 B3NQ41 B4GCG5 A0A3B0IZ35 B3MCP3 A0A3B0JIX6 A0A3B0IYV1 B4PAB8 B4J680 B4I8Y3 A0A336M6D5 A0A336KH44 A0A146L3R1 A0A3R7MSA7 A0A0L8I8M4 A0A1I8NG42 B4MRL5 B4KLY2 A0A1I8P1Z1 B4LP09 V4CRB5 A0A2H8TG57 A0A0M4EIL6 A0A0L0BQL8 T1IXP7 A0A0P4VYU1 A0A0P4W136 A0A0A1X7P6 A0A1B0CUP5 A0A0K8U519 A0A0K8UK47 A0A087TDE9 W8C8Q2 A0A2C9KE64 V9KFN7 W4YRS9 A0A2L2YGV9 A0A1A9ZZH8 A0A1A9V697 A0A1B0BW34 A0A2B4T1Z3 A0A0K2TV75 A0A1A9X9L8 A0A1A9X5T1 A0A293LQV3 A0A0T6BCR5 A0A099YR40 A0A2R5LDU8 M7ATY9 A0A091NHU4
PDB
5KHR
E-value=2.29587e-140,
Score=1280
Ontologies
GO
Topology
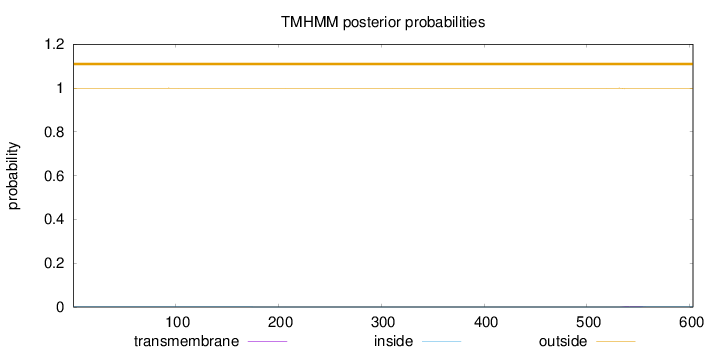
Length:
603
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06189
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00064
outside
1 - 603
Population Genetic Test Statistics
Pi
226.499648
Theta
212.181554
Tajima's D
0.282449
CLR
25.942932
CSRT
0.447827608619569
Interpretation
Uncertain
