Pre Gene Modal
BGIBMGA005151
Annotation
PREDICTED:_DNA_replication_licensing_factor_mcm5_[Papilio_polytes]
Full name
DNA helicase
+ More
DNA replication licensing factor Mcm5
DNA replication licensing factor mcm5
DNA replication licensing factor mcm5-A
DNA replication licensing factor mcm5-B
DNA replication licensing factor Mcm5
DNA replication licensing factor mcm5
DNA replication licensing factor mcm5-A
DNA replication licensing factor mcm5-B
Alternative Name
Minichromosome maintenance 5 protein
CDC46 homolog A
CDC46p
p92
CDC46 homolog B
CDC46 homolog A
CDC46p
p92
CDC46 homolog B
Location in the cell
Mitochondrial Reliability : 1.274 Nuclear Reliability : 1.717
Sequence
CDS
ATGGAAGGTTTTGATGATCCTGGTGTATTTTTCTCTGATAATTTTGGTGTAGAAGAAAACGAATCTCAAGATCATGTTAATTTACAAGCAGTCAAAAAGAAATTCAAAGAATTCATGAGACAGTTTCATACTGGAAACTTCAATTACAAATACAGAGATGCCCTCAAACGCAACTATAACCTAAACCAATATTGGGTCGAGATAAATATTGAAGATTTGTCTAGTTTTGATGACGTTCTTGCTGAAAAATTATACAAGAAACCAACTGAACATTTGCCTATACTAGAAGAAGCTGCTAAAGAGTTAGCTGATGAGTTGACTGCACCGAGACCAGAAGGTGAGGAGAAAGTTGAGGATATTCAAGTGCTTTTGTCATCAGATGCACATTCATCAAATTTGAGGGAGCTTAAATCAGAAACTGTATCAAGACTAGTGAAGATTCCTGGTATTGTAATATCAGCATCCGGTATTAAAGCAAAAGCCACAAAAATATCCATACAATGTCGCTCATGCCGCAATGTTATTCCCAATCTTCCTGTGAAGCCAGGTCTGGAAGGATATGTCATGCCAAGAAAATGCAATACAGAACAATCTGGACGTCCTAAATGTCCTTTAGATCCATATTTCATTATTCCCGACAAATGCAAATGTATCGATTATCAGGTGTTGAAACTTCAAGAAGCACCAGAAAGTATTCCACAAGGAGAAATGCCAAGGCATTTGACTGTATACTGTGATCGTACATTATGCGAACGTGTTGCACCAGGCGCACGAGTCACGGTTTTGGGGATTTATTCTATAAAAAAGATAGCAAAAGTAGGGCGCGAGGGTCGCGACAAAGGCTCCGTAGGCGTCCGGTCATCGTACCTACGAGCCGTAGGGCTCACCGCAGAGGAAGGTGTCACTAGCGGCCTGAGACCCTTTACTGCAGAAGAAGAAGAGCAATTCAGAAGACTCGCTGCCTCTCCAGACATTTATGAAAGGATCGGAAAGTCCATTGCTCCAAGTGTTTTTGGTGCTGTTGATATGAAGAAAGCTATCGCTTGTCTGCTTTTTGGAGGATCTAGAAAACGCCTACCGGATGGCCTGACTCGTAGAGGAGATATAAATATACTTTTACTTGGAGACCCTGGTACTGCCAAATCTCAATTGCTAAAATTTGTGGAGAAGGTTGCCCCGATTGGCGTTTATACCTCAGGGAAAGGTTCGAGTGCAGCAGGTCTCACCGCTTCAGTCATTAGAGATCCTGGAAGCCGTAACTTCGTAATGGAGGGAGGGGCTATGGTTTTGGCTGATGGTGGTGTTGTTTGTATAGACGAATTTGATAAGATGAGAGAAGATGATCGCGTCGCCATACACGAGGCAATGGAACAACAAACTATTTCTATTGCTAAGGCGGGTATCACCACTACATTAAATTCTCGTTGTTCTGTCCTGGCGGCCGCCAACTCTGTATTCGGACGATGGGACGATACTAAAGCTGATGATAACATTGATTTCATGCCAACAATTTTATCCAGATTCGACATGATTTTTATAGTCAAAGATGAACATGATCAGAACAGAGATATAACGCTCGCGAAGCACATAATCAGCGTGCACATGGGCGGAGAGAGCAGCGAGCGTGCGGCGGCGGCGGGCGAGCTGCCGCTGACGACGCTGCGTCGGTACAGCGCGTACTGCCGCGCGCGCTGCGGGCCCCGCCTGGCGCCGCCCGCCGCCGCCCGCCTGCAGGCGCGCTACGTGCTCATGCGGACCGGCGCGCTGCAACACGAGCGACAAGCCGACAAGCGTCTCTCCATACCCATCACCGTTAGGCAATTGGAAGCCATAGTTCGTATATCAGAATCTCTCGCTAAGATGCAGTTGCAACCGTTTGCGACTGAAGCTCACGTAAATGAAGCACTTCGTCTGTTCCAAGTGTCCACGCTAGATGCGGCTATGACTGGAAGTTTGGCAGGAGCTGAAGGTTTTACAACAGAAGAAGATCACGAAATGCTGACTCGCGTCGAGAAACAATTAAAGCGCAGGTTCGCTGTGGGCTCTCAGGTTTCTGAACAGACAATCATTCAAGACTTCTTGCGACAAAAATATCCAGAAAGAGCCATTCTTAAAGTAATACATATGATGATACGACGAGGGGAGCTTCAGCATAGATTACAGCGAAAAATGTTGTATAGACTCTCATAA
Protein
MEGFDDPGVFFSDNFGVEENESQDHVNLQAVKKKFKEFMRQFHTGNFNYKYRDALKRNYNLNQYWVEINIEDLSSFDDVLAEKLYKKPTEHLPILEEAAKELADELTAPRPEGEEKVEDIQVLLSSDAHSSNLRELKSETVSRLVKIPGIVISASGIKAKATKISIQCRSCRNVIPNLPVKPGLEGYVMPRKCNTEQSGRPKCPLDPYFIIPDKCKCIDYQVLKLQEAPESIPQGEMPRHLTVYCDRTLCERVAPGARVTVLGIYSIKKIAKVGREGRDKGSVGVRSSYLRAVGLTAEEGVTSGLRPFTAEEEEQFRRLAASPDIYERIGKSIAPSVFGAVDMKKAIACLLFGGSRKRLPDGLTRRGDINILLLGDPGTAKSQLLKFVEKVAPIGVYTSGKGSSAAGLTASVIRDPGSRNFVMEGGAMVLADGGVVCIDEFDKMREDDRVAIHEAMEQQTISIAKAGITTTLNSRCSVLAAANSVFGRWDDTKADDNIDFMPTILSRFDMIFIVKDEHDQNRDITLAKHIISVHMGGESSERAAAAGELPLTTLRRYSAYCRARCGPRLAPPAAARLQARYVLMRTGALQHERQADKRLSIPITVRQLEAIVRISESLAKMQLQPFATEAHVNEALRLFQVSTLDAAMTGSLAGAEGFTTEEDHEMLTRVEKQLKRRFAVGSQVSEQTIIQDFLRQKYPERAILKVIHMMIRRGELQHRLQRKMLYRLS
Summary
Description
Acts as component of the Mcm2-7 complex (Mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the Mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity.
Acts as component of the mcm2-7 complex (mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity (By similarity).
Acts as component of the mcm2-7 complex (mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity (By similarity).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Component of the Mcm2-7 complex. The complex forms a toroidal hexameric ring with the proposed subunit order Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5 (Probable).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (By simililarity).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2.
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5. The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2 (By similarity).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (By simililarity).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2.
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5. The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2 (By similarity).
Similarity
Belongs to the MCM family.
Keywords
ATP-binding
Cell cycle
Complete proteome
Cytoplasm
DNA replication
DNA-binding
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Reference proteome
Alternative splicing
Feature
chain DNA replication licensing factor Mcm5
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2H1W380
A0A2W1BKQ9
H9J6K9
A0A194PI49
A0A212F8Y1
A0A2P8Z223
+ More
A0A2J7PYT0 A0A067QM81 A0A1B6D033 A0A0N0BDP3 A0A194RLA6 A0A3R7QW06 A0A154PHC8 A0A0J7NTI1 A0A2A3ERL1 A0A088A0G3 Q17H38 A0A182GQF3 A0A182FAF4 A0A026WH14 E2BWT0 E2A7N9 B0WNG2 W5JTL1 A0A182NCU9 A0A182PLH0 A0A089GK37 A0A151X597 A0A182M566 A0A182RI25 A0A158NAF4 A0A182W384 E9H6I5 A0A182VKP3 A0A084VDK0 A0A182XF67 A0A164K478 A0A1W4WBT4 A0A182Y908 A0A182K4W3 A0A1Q3EVG0 A0A182UHP0 A0A182KZR5 Q7QA70 A0A182HVN8 A0A182QVG1 A0A0P5V2S2 A0A0P6JBI5 A0A182JCU9 A0A0P5HSI7 A0A2P2HWJ7 A0A0C9RED7 A0A336L2Y3 A0A224YX91 A0A195CKR6 A0A0A1X8M6 A0A0C9S360 A0A131YXL0 L7M9Q6 A0A1Y1MI36 A0A0L7R1I7 W8BKX8 B3P1K0 B4QUP5 A0A034WVN9 B4HIQ0 Q9VGW6 B4PLD4 B3M291 A0A1E1X2S1 B4NJY3 B4KC98 A0A0L0BQ18 A0A1I8NCC4 A0A0P4W8T1 U5ELZ4 A0A0M5J5H1 A0A1W4W052 A0A1Z5L9M3 R7V2U4 A0A210Q0K1 A0A3B0JQH7 Q561P5 A0A1I8Q5I2 B4JHM4 A0A2R5LIX8 A0A293LWJ3 A0A1L8GNF3 P55862 K7J409 A0A099ZFS6 A0A091HGL6 A0A2I0USR1 Q5ZKL0 A0A232FEX0 A0A131XFY4 V5HPK2 A0A1L8GGS4 B4M5I3 Q6PCI7
A0A2J7PYT0 A0A067QM81 A0A1B6D033 A0A0N0BDP3 A0A194RLA6 A0A3R7QW06 A0A154PHC8 A0A0J7NTI1 A0A2A3ERL1 A0A088A0G3 Q17H38 A0A182GQF3 A0A182FAF4 A0A026WH14 E2BWT0 E2A7N9 B0WNG2 W5JTL1 A0A182NCU9 A0A182PLH0 A0A089GK37 A0A151X597 A0A182M566 A0A182RI25 A0A158NAF4 A0A182W384 E9H6I5 A0A182VKP3 A0A084VDK0 A0A182XF67 A0A164K478 A0A1W4WBT4 A0A182Y908 A0A182K4W3 A0A1Q3EVG0 A0A182UHP0 A0A182KZR5 Q7QA70 A0A182HVN8 A0A182QVG1 A0A0P5V2S2 A0A0P6JBI5 A0A182JCU9 A0A0P5HSI7 A0A2P2HWJ7 A0A0C9RED7 A0A336L2Y3 A0A224YX91 A0A195CKR6 A0A0A1X8M6 A0A0C9S360 A0A131YXL0 L7M9Q6 A0A1Y1MI36 A0A0L7R1I7 W8BKX8 B3P1K0 B4QUP5 A0A034WVN9 B4HIQ0 Q9VGW6 B4PLD4 B3M291 A0A1E1X2S1 B4NJY3 B4KC98 A0A0L0BQ18 A0A1I8NCC4 A0A0P4W8T1 U5ELZ4 A0A0M5J5H1 A0A1W4W052 A0A1Z5L9M3 R7V2U4 A0A210Q0K1 A0A3B0JQH7 Q561P5 A0A1I8Q5I2 B4JHM4 A0A2R5LIX8 A0A293LWJ3 A0A1L8GNF3 P55862 K7J409 A0A099ZFS6 A0A091HGL6 A0A2I0USR1 Q5ZKL0 A0A232FEX0 A0A131XFY4 V5HPK2 A0A1L8GGS4 B4M5I3 Q6PCI7
EC Number
3.6.4.12
Pubmed
28756777
19121390
26354079
22118469
29403074
24845553
+ More
17510324 26483478 24508170 30249741 20798317 20920257 23761445 21347285 21292972 24438588 25244985 20966253 12364791 14747013 17210077 28797301 25830018 26131772 26830274 25576852 28004739 24495485 17994087 25348373 9224901 10731132 12537572 16798881 20122406 17550304 28503490 26108605 25315136 28528879 23254933 28812685 27762356 9214647 8917078 9214646 9851868 16369567 20075255 15592404 15642098 28648823 28049606 25765539
17510324 26483478 24508170 30249741 20798317 20920257 23761445 21347285 21292972 24438588 25244985 20966253 12364791 14747013 17210077 28797301 25830018 26131772 26830274 25576852 28004739 24495485 17994087 25348373 9224901 10731132 12537572 16798881 20122406 17550304 28503490 26108605 25315136 28528879 23254933 28812685 27762356 9214647 8917078 9214646 9851868 16369567 20075255 15592404 15642098 28648823 28049606 25765539
EMBL
ODYU01006049
SOQ47560.1
KZ149994
PZC75478.1
BABH01019865
BABH01019866
+ More
KQ459604 KPI92404.1 AGBW02009658 OWR50187.1 PYGN01000235 PSN50546.1 NEVH01020342 PNF21485.1 KK853179 KDR10221.1 GEDC01018365 JAS18933.1 KQ435863 KOX70391.1 KQ460045 KPJ18100.1 QCYY01001128 ROT80337.1 KQ434905 KZC11207.1 LBMM01001798 KMQ95730.1 KZ288194 PBC33912.1 CH477253 EAT45976.1 JXUM01080405 JXUM01094528 KQ564135 KQ563183 KXJ72707.1 KXJ74319.1 KK107231 QOIP01000009 EZA54966.1 RLU18576.1 GL451188 EFN79794.1 GL437365 EFN70560.1 DS232010 EDS31616.1 ADMH02000180 ETN67461.1 KM036513 AIP98400.1 KQ982519 KYQ55494.1 AXCM01000104 ADTU01010273 GL732597 EFX72666.1 ATLV01011556 KE524695 KFB36044.1 LRGB01003375 KZS02936.1 GFDL01015744 JAV19301.1 AAAB01008898 EAA09249.2 APCN01005808 AXCN02001206 GDIP01104936 GDIQ01019924 JAL98778.1 JAN74813.1 GDIQ01019925 JAN74812.1 GDIQ01229345 JAK22380.1 IACF01000398 LAB66175.1 GBYB01013200 GBYB01014969 JAG82967.1 JAG84736.1 UFQS01001768 UFQT01001768 SSX12278.1 SSX31729.1 GFPF01007296 MAA18442.1 KQ977642 KYN01047.1 GBXI01007202 JAD07090.1 GBZX01001246 JAG91494.1 GEDV01005342 JAP83215.1 GACK01005146 JAA59888.1 GEZM01036149 JAV82937.1 KQ414667 KOC64709.1 GAMC01008922 JAB97633.1 CH954181 EDV49599.1 CM000364 EDX13452.1 GAKP01001104 JAC57848.1 CH480815 EDW42697.1 U83493 AE014297 AY071628 CM000160 EDW96841.1 CH902617 EDV42282.1 GFAC01005623 JAT93565.1 CH964272 EDW83985.1 CH933806 EDW13707.1 JRES01001539 KNC22180.1 GDRN01063226 JAI64995.1 GANO01001217 JAB58654.1 CP012526 ALC47711.1 GFJQ02002864 JAW04106.1 AMQN01000770 KB295623 ELU12874.1 NEDP02005311 OWF42189.1 OUUW01000007 SPP83213.1 CR848584 BC093455 AAH93455.1 CAJ81496.1 CH916369 EDV93863.1 GGLE01005303 MBY09429.1 GFWV01007408 MAA32138.1 CM004472 OCT85375.1 U44048 D63920 BC047250 AAZX01001005 KL892383 KGL79685.1 KL217466 KFO95403.1 KZ505643 PKU49093.1 AC145933 AJ720074 CAG31733.1 NNAY01000320 OXU29242.1 GEFH01003960 JAP64621.1 GANP01006826 JAB77642.1 CM004473 OCT83020.1 CH940652 EDW58909.1 BC059310 AAH59310.1
KQ459604 KPI92404.1 AGBW02009658 OWR50187.1 PYGN01000235 PSN50546.1 NEVH01020342 PNF21485.1 KK853179 KDR10221.1 GEDC01018365 JAS18933.1 KQ435863 KOX70391.1 KQ460045 KPJ18100.1 QCYY01001128 ROT80337.1 KQ434905 KZC11207.1 LBMM01001798 KMQ95730.1 KZ288194 PBC33912.1 CH477253 EAT45976.1 JXUM01080405 JXUM01094528 KQ564135 KQ563183 KXJ72707.1 KXJ74319.1 KK107231 QOIP01000009 EZA54966.1 RLU18576.1 GL451188 EFN79794.1 GL437365 EFN70560.1 DS232010 EDS31616.1 ADMH02000180 ETN67461.1 KM036513 AIP98400.1 KQ982519 KYQ55494.1 AXCM01000104 ADTU01010273 GL732597 EFX72666.1 ATLV01011556 KE524695 KFB36044.1 LRGB01003375 KZS02936.1 GFDL01015744 JAV19301.1 AAAB01008898 EAA09249.2 APCN01005808 AXCN02001206 GDIP01104936 GDIQ01019924 JAL98778.1 JAN74813.1 GDIQ01019925 JAN74812.1 GDIQ01229345 JAK22380.1 IACF01000398 LAB66175.1 GBYB01013200 GBYB01014969 JAG82967.1 JAG84736.1 UFQS01001768 UFQT01001768 SSX12278.1 SSX31729.1 GFPF01007296 MAA18442.1 KQ977642 KYN01047.1 GBXI01007202 JAD07090.1 GBZX01001246 JAG91494.1 GEDV01005342 JAP83215.1 GACK01005146 JAA59888.1 GEZM01036149 JAV82937.1 KQ414667 KOC64709.1 GAMC01008922 JAB97633.1 CH954181 EDV49599.1 CM000364 EDX13452.1 GAKP01001104 JAC57848.1 CH480815 EDW42697.1 U83493 AE014297 AY071628 CM000160 EDW96841.1 CH902617 EDV42282.1 GFAC01005623 JAT93565.1 CH964272 EDW83985.1 CH933806 EDW13707.1 JRES01001539 KNC22180.1 GDRN01063226 JAI64995.1 GANO01001217 JAB58654.1 CP012526 ALC47711.1 GFJQ02002864 JAW04106.1 AMQN01000770 KB295623 ELU12874.1 NEDP02005311 OWF42189.1 OUUW01000007 SPP83213.1 CR848584 BC093455 AAH93455.1 CAJ81496.1 CH916369 EDV93863.1 GGLE01005303 MBY09429.1 GFWV01007408 MAA32138.1 CM004472 OCT85375.1 U44048 D63920 BC047250 AAZX01001005 KL892383 KGL79685.1 KL217466 KFO95403.1 KZ505643 PKU49093.1 AC145933 AJ720074 CAG31733.1 NNAY01000320 OXU29242.1 GEFH01003960 JAP64621.1 GANP01006826 JAB77642.1 CM004473 OCT83020.1 CH940652 EDW58909.1 BC059310 AAH59310.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000245037
UP000235965
UP000027135
+ More
UP000053105 UP000053240 UP000283509 UP000076502 UP000036403 UP000242457 UP000005203 UP000008820 UP000069940 UP000249989 UP000069272 UP000053097 UP000279307 UP000008237 UP000000311 UP000002320 UP000000673 UP000075884 UP000075885 UP000075809 UP000075883 UP000075900 UP000005205 UP000075920 UP000000305 UP000075903 UP000030765 UP000076407 UP000076858 UP000192223 UP000076408 UP000075881 UP000075902 UP000075882 UP000007062 UP000075840 UP000075886 UP000075880 UP000078542 UP000053825 UP000008711 UP000000304 UP000001292 UP000000803 UP000002282 UP000007801 UP000007798 UP000009192 UP000037069 UP000095301 UP000092553 UP000192221 UP000014760 UP000242188 UP000268350 UP000008143 UP000095300 UP000001070 UP000186698 UP000002358 UP000053641 UP000054308 UP000000539 UP000215335 UP000008792
UP000053105 UP000053240 UP000283509 UP000076502 UP000036403 UP000242457 UP000005203 UP000008820 UP000069940 UP000249989 UP000069272 UP000053097 UP000279307 UP000008237 UP000000311 UP000002320 UP000000673 UP000075884 UP000075885 UP000075809 UP000075883 UP000075900 UP000005205 UP000075920 UP000000305 UP000075903 UP000030765 UP000076407 UP000076858 UP000192223 UP000076408 UP000075881 UP000075902 UP000075882 UP000007062 UP000075840 UP000075886 UP000075880 UP000078542 UP000053825 UP000008711 UP000000304 UP000001292 UP000000803 UP000002282 UP000007801 UP000007798 UP000009192 UP000037069 UP000095301 UP000092553 UP000192221 UP000014760 UP000242188 UP000268350 UP000008143 UP000095300 UP000001070 UP000186698 UP000002358 UP000053641 UP000054308 UP000000539 UP000215335 UP000008792
Interpro
ProteinModelPortal
A0A2H1W380
A0A2W1BKQ9
H9J6K9
A0A194PI49
A0A212F8Y1
A0A2P8Z223
+ More
A0A2J7PYT0 A0A067QM81 A0A1B6D033 A0A0N0BDP3 A0A194RLA6 A0A3R7QW06 A0A154PHC8 A0A0J7NTI1 A0A2A3ERL1 A0A088A0G3 Q17H38 A0A182GQF3 A0A182FAF4 A0A026WH14 E2BWT0 E2A7N9 B0WNG2 W5JTL1 A0A182NCU9 A0A182PLH0 A0A089GK37 A0A151X597 A0A182M566 A0A182RI25 A0A158NAF4 A0A182W384 E9H6I5 A0A182VKP3 A0A084VDK0 A0A182XF67 A0A164K478 A0A1W4WBT4 A0A182Y908 A0A182K4W3 A0A1Q3EVG0 A0A182UHP0 A0A182KZR5 Q7QA70 A0A182HVN8 A0A182QVG1 A0A0P5V2S2 A0A0P6JBI5 A0A182JCU9 A0A0P5HSI7 A0A2P2HWJ7 A0A0C9RED7 A0A336L2Y3 A0A224YX91 A0A195CKR6 A0A0A1X8M6 A0A0C9S360 A0A131YXL0 L7M9Q6 A0A1Y1MI36 A0A0L7R1I7 W8BKX8 B3P1K0 B4QUP5 A0A034WVN9 B4HIQ0 Q9VGW6 B4PLD4 B3M291 A0A1E1X2S1 B4NJY3 B4KC98 A0A0L0BQ18 A0A1I8NCC4 A0A0P4W8T1 U5ELZ4 A0A0M5J5H1 A0A1W4W052 A0A1Z5L9M3 R7V2U4 A0A210Q0K1 A0A3B0JQH7 Q561P5 A0A1I8Q5I2 B4JHM4 A0A2R5LIX8 A0A293LWJ3 A0A1L8GNF3 P55862 K7J409 A0A099ZFS6 A0A091HGL6 A0A2I0USR1 Q5ZKL0 A0A232FEX0 A0A131XFY4 V5HPK2 A0A1L8GGS4 B4M5I3 Q6PCI7
A0A2J7PYT0 A0A067QM81 A0A1B6D033 A0A0N0BDP3 A0A194RLA6 A0A3R7QW06 A0A154PHC8 A0A0J7NTI1 A0A2A3ERL1 A0A088A0G3 Q17H38 A0A182GQF3 A0A182FAF4 A0A026WH14 E2BWT0 E2A7N9 B0WNG2 W5JTL1 A0A182NCU9 A0A182PLH0 A0A089GK37 A0A151X597 A0A182M566 A0A182RI25 A0A158NAF4 A0A182W384 E9H6I5 A0A182VKP3 A0A084VDK0 A0A182XF67 A0A164K478 A0A1W4WBT4 A0A182Y908 A0A182K4W3 A0A1Q3EVG0 A0A182UHP0 A0A182KZR5 Q7QA70 A0A182HVN8 A0A182QVG1 A0A0P5V2S2 A0A0P6JBI5 A0A182JCU9 A0A0P5HSI7 A0A2P2HWJ7 A0A0C9RED7 A0A336L2Y3 A0A224YX91 A0A195CKR6 A0A0A1X8M6 A0A0C9S360 A0A131YXL0 L7M9Q6 A0A1Y1MI36 A0A0L7R1I7 W8BKX8 B3P1K0 B4QUP5 A0A034WVN9 B4HIQ0 Q9VGW6 B4PLD4 B3M291 A0A1E1X2S1 B4NJY3 B4KC98 A0A0L0BQ18 A0A1I8NCC4 A0A0P4W8T1 U5ELZ4 A0A0M5J5H1 A0A1W4W052 A0A1Z5L9M3 R7V2U4 A0A210Q0K1 A0A3B0JQH7 Q561P5 A0A1I8Q5I2 B4JHM4 A0A2R5LIX8 A0A293LWJ3 A0A1L8GNF3 P55862 K7J409 A0A099ZFS6 A0A091HGL6 A0A2I0USR1 Q5ZKL0 A0A232FEX0 A0A131XFY4 V5HPK2 A0A1L8GGS4 B4M5I3 Q6PCI7
PDB
6HV9
E-value=8.4023e-164,
Score=1483
Ontologies
GO
GO:0005634
GO:0006270
GO:0005524
GO:0004386
GO:0042555
GO:0003688
GO:0003677
GO:0006260
GO:0000727
GO:0032508
GO:0006267
GO:0003697
GO:0042023
GO:0043138
GO:0000712
GO:0030261
GO:0005829
GO:0051321
GO:0000278
GO:0043066
GO:0030174
GO:0006268
GO:0043009
GO:0000785
GO:0007049
GO:0005654
GO:0003678
GO:0003682
GO:0015930
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Cytosol
Cytoplasm
Cytosol
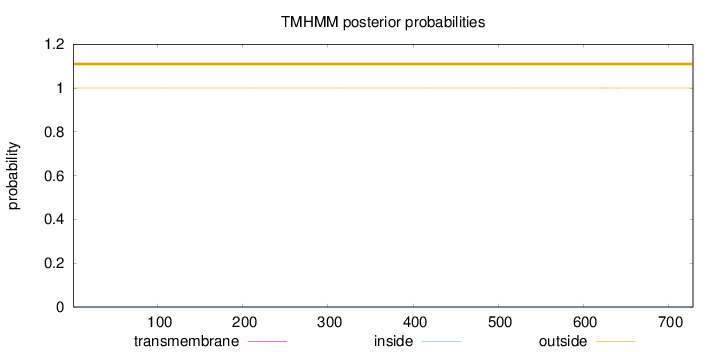
Length:
729
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00321
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 729
Population Genetic Test Statistics
Pi
255.7237
Theta
190.060606
Tajima's D
0.721308
CLR
0.212317
CSRT
0.579821008949553
Interpretation
Uncertain
