Pre Gene Modal
BGIBMGA005047
Annotation
PREDICTED:_1?5-anhydro-D-fructose_reductase-like_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 3.421
Sequence
CDS
ATGAAGCACGTGGAACTTTCATATACGAAAGACTTGATGCCAACATTAGGTCTTGGAACGTGGCAAGCTCCCTCTGATGTTATAGAGTCTACTGTATACAAAGCCTTAGATTTGGGCTATAGGCACATAGATACAGCATTTAATTATAATAATGAGGAATCTATCGGAAGAGCCCTTAAGAAGTGGATTGATGATGGCAAAGGAACAAGAACTGATTTATTTGTCACGACTAAGCTTCCTCACGTGGGTAATCGCGCATCTGATGTGAAGAAATTTTTAGAACTCCAGCTTAGTCGACTGCAGTTGGACTATGTCGATTTATATTTAATTCACGTTCCGTTCGGGTTCCACTGCGATCCGGAGACGCTGACTCCAAGGGTCAAGAGTGATGGGGAATACGACGTAGATATGGATACCAATCATCTTACTATTTGGAAGGAAATGGAACTATGTCAAAGGGAAGGTTTAATCCGTAATCTTGGTTTGTCGAATTTCAACGAGGAGCAGATAAGAAAAATTTCTAAGTCATCTACAATCAAGCCACAAGTCCTCCAAGTAGAATTGCATGCGTATTTCCAACAACAGGGACTTCGGAAGTTTTGTACCGATAATGACATCGTAGTCACGGCTTACGCGCCTTTGGGTAGTCCCGGAGCTAAAGATCATTTCGTTACCAAATACAACTATAGCCCGGATACGTTCCCCGATTTACTAAACTTACCACCAGTAAAAGAGATTGCTGATAAGCACGGCCGAACACCAGCACAAATATTACTAAACTTCTTAGTACAACAAAAGGTCGTTGTAATACCAAAGAGCACAAGTGAACATAGACTAAGAGAAAACATGAATATTTACGATTTCGAGTTGTCAGCGCCTGAAATGGATTTGCTAAAGAAGTTGGACAAGGGGGAGAGTGGTCGTATTTTTAATTTTCTATTCTGGAAAGGTGTGGAGAAACACCCAGAATATCCGTTCCATATTGAAAAAATTGATTCCAGCAAAGACAAACATTAA
Protein
MKHVELSYTKDLMPTLGLGTWQAPSDVIESTVYKALDLGYRHIDTAFNYNNEESIGRALKKWIDDGKGTRTDLFVTTKLPHVGNRASDVKKFLELQLSRLQLDYVDLYLIHVPFGFHCDPETLTPRVKSDGEYDVDMDTNHLTIWKEMELCQREGLIRNLGLSNFNEEQIRKISKSSTIKPQVLQVELHAYFQQQGLRKFCTDNDIVVTAYAPLGSPGAKDHFVTKYNYSPDTFPDLLNLPPVKEIADKHGRTPAQILLNFLVQQKVVVIPKSTSEHRLRENMNIYDFELSAPEMDLLKKLDKGESGRIFNFLFWKGVEKHPEYPFHIEKIDSSKDKH
Summary
Uniprot
H9J6A6
A0A194RPZ9
A0A194PH75
A0A3S2NNA8
A0A0F6Q2L9
A0A291P0X4
+ More
A0A2W1BKB2 A0A291P0T8 A0A212F8V9 A0A2H1W3A4 K7J405 A0A0L7R1J1 D6WTS5 A0A026WFS5 A0A195CK08 A0A158NAF6 A0A195EVL4 A0A2J7Q2J5 A0A232EVU0 A0A2A3ES93 A0A087ZZR5 A0A0N0BDG0 A0A067REM7 E2A7P0 A0A151X4Z0 A0A154PIR2 E2BWS9 E0VSM5 A0A0J7KZD8 J3JZ53 U4U7N8 N6TT69 F4WMJ5 K7IP18 A0A0L7LW80 A0A232F739 A0A232EUU6 K7INV3 A0A1B6CPV8 A0A151I118 K7J407 A0A0P6FC08 A0A140CVV2 A0A336KNX9 A0A164ZMS8 A0A310S6U0 A0A182QB38 A0A2J7QCW6 E9GI56 A0A0A9YK38 D6WTU2 A0A1A9XM90 A0A182NKU4 A0A0N8DLF9 A0A182V201 A0A164KJP0 A0A182XK76 A0A0P6K0A8 A0A0P5DG11 Q7QDK4 A0A182HJR2 A0A0N8DQI7 A0A0P5DMC5 A0A182M2M9 A0A1B0BNU1 A0A164ZMP8 A0A0N8AMB7 A0A182W0L0 A0A182KWY7 A0A182R804 E2BWT3 A0A0P5BK85 B4MBK5 A0A1Y1MCE5 A0A0P4ZBC9 A0A0N8AEZ3 A0A026WFT0 A0A0P5YA29 A0A0P5M6K1 A0A0P5FT22 A0A0P5FM74 A0A0P6JPS0 A0A195CLU3 B3NYU4 A0A182IP00 A0A182Y4I9 A0A1B0FNR1 A0A0P6ASR2 B0W9B1 A0A1B2M4X3 A0A1A9V8U1 A0A1I8Q3N3 A0A0P5V7I4 A0A2J7QZP5 A0A0N8ACN7 A0A087ZZ66 A0A067QPZ1 D3TRK6 A0A232FFF2
A0A2W1BKB2 A0A291P0T8 A0A212F8V9 A0A2H1W3A4 K7J405 A0A0L7R1J1 D6WTS5 A0A026WFS5 A0A195CK08 A0A158NAF6 A0A195EVL4 A0A2J7Q2J5 A0A232EVU0 A0A2A3ES93 A0A087ZZR5 A0A0N0BDG0 A0A067REM7 E2A7P0 A0A151X4Z0 A0A154PIR2 E2BWS9 E0VSM5 A0A0J7KZD8 J3JZ53 U4U7N8 N6TT69 F4WMJ5 K7IP18 A0A0L7LW80 A0A232F739 A0A232EUU6 K7INV3 A0A1B6CPV8 A0A151I118 K7J407 A0A0P6FC08 A0A140CVV2 A0A336KNX9 A0A164ZMS8 A0A310S6U0 A0A182QB38 A0A2J7QCW6 E9GI56 A0A0A9YK38 D6WTU2 A0A1A9XM90 A0A182NKU4 A0A0N8DLF9 A0A182V201 A0A164KJP0 A0A182XK76 A0A0P6K0A8 A0A0P5DG11 Q7QDK4 A0A182HJR2 A0A0N8DQI7 A0A0P5DMC5 A0A182M2M9 A0A1B0BNU1 A0A164ZMP8 A0A0N8AMB7 A0A182W0L0 A0A182KWY7 A0A182R804 E2BWT3 A0A0P5BK85 B4MBK5 A0A1Y1MCE5 A0A0P4ZBC9 A0A0N8AEZ3 A0A026WFT0 A0A0P5YA29 A0A0P5M6K1 A0A0P5FT22 A0A0P5FM74 A0A0P6JPS0 A0A195CLU3 B3NYU4 A0A182IP00 A0A182Y4I9 A0A1B0FNR1 A0A0P6ASR2 B0W9B1 A0A1B2M4X3 A0A1A9V8U1 A0A1I8Q3N3 A0A0P5V7I4 A0A2J7QZP5 A0A0N8ACN7 A0A087ZZ66 A0A067QPZ1 D3TRK6 A0A232FFF2
Pubmed
EMBL
BABH01019866
KQ460045
KPJ18101.1
KQ459604
KPI92403.1
RSAL01000001
+ More
RVE55316.1 KP237898 AKD01751.1 MF687616 ATJ44542.1 KZ149994 PZC75479.1 MF687577 ATJ44503.1 AGBW02009658 OWR50186.1 ODYU01006049 SOQ47561.1 AAZX01001010 KQ414667 KOC64708.1 KQ971352 EFA06738.2 KK107231 QOIP01000009 EZA54965.1 RLU18577.1 KQ977642 KYN01048.1 ADTU01010273 KQ981954 KYN32181.1 NEVH01019370 PNF22802.1 NNAY01001957 OXU22457.1 KZ288194 PBC33911.1 KQ435863 KOX70392.1 KK852683 KDR18534.1 GL437365 EFN70561.1 KQ982519 KYQ55495.1 KQ434905 KZC11208.1 GL451188 EFN79793.1 DS235751 EEB16381.1 LBMM01001798 KMQ95731.1 BT128534 AEE63491.1 KB632194 ERL89909.1 APGK01055580 KB741266 ENN71536.1 GL888217 EGI64606.1 JTDY01000014 KOB79416.1 NNAY01000838 OXU26253.1 NNAY01002091 OXU22106.1 GEDC01021789 JAS15509.1 KQ976600 KYM79469.1 GDIQ01062257 JAN32480.1 KU686221 AMJ21949.2 UFQS01000309 UFQT01000309 SSX02710.1 SSX23084.1 LRGB01000704 KZS16526.1 KQ767242 OAD53474.1 AXCN02000306 NEVH01015858 PNF26431.1 GL732546 EFX80617.1 GBHO01011065 JAG32539.1 EFA07327.1 GDIP01022175 JAM81540.1 LRGB01003310 KZS03322.1 GDIQ01000679 JAN94058.1 GDIP01158000 JAJ65402.1 AAAB01008851 EAA07379.5 APCN01002207 GDIP01010751 JAM92964.1 GDIP01153991 JAJ69411.1 AXCM01000236 JXJN01017597 KZS16524.1 GDIQ01261723 JAJ90001.1 EFN79797.1 GDIP01183847 JAJ39555.1 CH940656 EDW58476.1 GEZM01038692 JAV81556.1 GDIP01218634 JAJ04768.1 GDIP01153993 JAJ69409.1 EZA54970.1 GDIP01074413 JAM29302.1 KZS16504.1 GDIQ01163890 JAK87835.1 GDIQ01258124 JAJ93600.1 GDIP01146455 JAJ76947.1 GDIQ01001498 JAN93239.1 KYN01044.1 CH954181 EDV48207.1 CCAG010016865 GDIP01024965 JAM78750.1 DS231863 EDS39971.1 KX447599 AOA60266.1 GDIP01103153 JAM00562.1 NEVH01009067 PNF34058.1 GDIP01160441 JAJ62961.1 KK853074 KDR11767.1 EZ424058 ADD20334.1 NNAY01000320 OXU29250.1
RVE55316.1 KP237898 AKD01751.1 MF687616 ATJ44542.1 KZ149994 PZC75479.1 MF687577 ATJ44503.1 AGBW02009658 OWR50186.1 ODYU01006049 SOQ47561.1 AAZX01001010 KQ414667 KOC64708.1 KQ971352 EFA06738.2 KK107231 QOIP01000009 EZA54965.1 RLU18577.1 KQ977642 KYN01048.1 ADTU01010273 KQ981954 KYN32181.1 NEVH01019370 PNF22802.1 NNAY01001957 OXU22457.1 KZ288194 PBC33911.1 KQ435863 KOX70392.1 KK852683 KDR18534.1 GL437365 EFN70561.1 KQ982519 KYQ55495.1 KQ434905 KZC11208.1 GL451188 EFN79793.1 DS235751 EEB16381.1 LBMM01001798 KMQ95731.1 BT128534 AEE63491.1 KB632194 ERL89909.1 APGK01055580 KB741266 ENN71536.1 GL888217 EGI64606.1 JTDY01000014 KOB79416.1 NNAY01000838 OXU26253.1 NNAY01002091 OXU22106.1 GEDC01021789 JAS15509.1 KQ976600 KYM79469.1 GDIQ01062257 JAN32480.1 KU686221 AMJ21949.2 UFQS01000309 UFQT01000309 SSX02710.1 SSX23084.1 LRGB01000704 KZS16526.1 KQ767242 OAD53474.1 AXCN02000306 NEVH01015858 PNF26431.1 GL732546 EFX80617.1 GBHO01011065 JAG32539.1 EFA07327.1 GDIP01022175 JAM81540.1 LRGB01003310 KZS03322.1 GDIQ01000679 JAN94058.1 GDIP01158000 JAJ65402.1 AAAB01008851 EAA07379.5 APCN01002207 GDIP01010751 JAM92964.1 GDIP01153991 JAJ69411.1 AXCM01000236 JXJN01017597 KZS16524.1 GDIQ01261723 JAJ90001.1 EFN79797.1 GDIP01183847 JAJ39555.1 CH940656 EDW58476.1 GEZM01038692 JAV81556.1 GDIP01218634 JAJ04768.1 GDIP01153993 JAJ69409.1 EZA54970.1 GDIP01074413 JAM29302.1 KZS16504.1 GDIQ01163890 JAK87835.1 GDIQ01258124 JAJ93600.1 GDIP01146455 JAJ76947.1 GDIQ01001498 JAN93239.1 KYN01044.1 CH954181 EDV48207.1 CCAG010016865 GDIP01024965 JAM78750.1 DS231863 EDS39971.1 KX447599 AOA60266.1 GDIP01103153 JAM00562.1 NEVH01009067 PNF34058.1 GDIP01160441 JAJ62961.1 KK853074 KDR11767.1 EZ424058 ADD20334.1 NNAY01000320 OXU29250.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000002358
+ More
UP000053825 UP000007266 UP000053097 UP000279307 UP000078542 UP000005205 UP000078541 UP000235965 UP000215335 UP000242457 UP000005203 UP000053105 UP000027135 UP000000311 UP000075809 UP000076502 UP000008237 UP000009046 UP000036403 UP000030742 UP000019118 UP000007755 UP000037510 UP000078540 UP000076858 UP000075886 UP000000305 UP000092443 UP000075884 UP000075903 UP000076407 UP000007062 UP000075840 UP000075883 UP000092460 UP000075920 UP000075882 UP000075900 UP000008792 UP000008711 UP000075880 UP000076408 UP000092444 UP000002320 UP000078200 UP000095300
UP000053825 UP000007266 UP000053097 UP000279307 UP000078542 UP000005205 UP000078541 UP000235965 UP000215335 UP000242457 UP000005203 UP000053105 UP000027135 UP000000311 UP000075809 UP000076502 UP000008237 UP000009046 UP000036403 UP000030742 UP000019118 UP000007755 UP000037510 UP000078540 UP000076858 UP000075886 UP000000305 UP000092443 UP000075884 UP000075903 UP000076407 UP000007062 UP000075840 UP000075883 UP000092460 UP000075920 UP000075882 UP000075900 UP000008792 UP000008711 UP000075880 UP000076408 UP000092444 UP000002320 UP000078200 UP000095300
PRIDE
Interpro
IPR023210
NADP_OxRdtase_dom
+ More
IPR036812 NADP_OxRdtase_dom_sf
IPR018170 Aldo/ket_reductase_CS
IPR020471 Aldo/keto_reductase
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR041715 HisRS-like_core
IPR000719 Prot_kinase_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR020472 G-protein_beta_WD-40_rep
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR036812 NADP_OxRdtase_dom_sf
IPR018170 Aldo/ket_reductase_CS
IPR020471 Aldo/keto_reductase
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR041715 HisRS-like_core
IPR000719 Prot_kinase_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR020472 G-protein_beta_WD-40_rep
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
Gene 3D
ProteinModelPortal
H9J6A6
A0A194RPZ9
A0A194PH75
A0A3S2NNA8
A0A0F6Q2L9
A0A291P0X4
+ More
A0A2W1BKB2 A0A291P0T8 A0A212F8V9 A0A2H1W3A4 K7J405 A0A0L7R1J1 D6WTS5 A0A026WFS5 A0A195CK08 A0A158NAF6 A0A195EVL4 A0A2J7Q2J5 A0A232EVU0 A0A2A3ES93 A0A087ZZR5 A0A0N0BDG0 A0A067REM7 E2A7P0 A0A151X4Z0 A0A154PIR2 E2BWS9 E0VSM5 A0A0J7KZD8 J3JZ53 U4U7N8 N6TT69 F4WMJ5 K7IP18 A0A0L7LW80 A0A232F739 A0A232EUU6 K7INV3 A0A1B6CPV8 A0A151I118 K7J407 A0A0P6FC08 A0A140CVV2 A0A336KNX9 A0A164ZMS8 A0A310S6U0 A0A182QB38 A0A2J7QCW6 E9GI56 A0A0A9YK38 D6WTU2 A0A1A9XM90 A0A182NKU4 A0A0N8DLF9 A0A182V201 A0A164KJP0 A0A182XK76 A0A0P6K0A8 A0A0P5DG11 Q7QDK4 A0A182HJR2 A0A0N8DQI7 A0A0P5DMC5 A0A182M2M9 A0A1B0BNU1 A0A164ZMP8 A0A0N8AMB7 A0A182W0L0 A0A182KWY7 A0A182R804 E2BWT3 A0A0P5BK85 B4MBK5 A0A1Y1MCE5 A0A0P4ZBC9 A0A0N8AEZ3 A0A026WFT0 A0A0P5YA29 A0A0P5M6K1 A0A0P5FT22 A0A0P5FM74 A0A0P6JPS0 A0A195CLU3 B3NYU4 A0A182IP00 A0A182Y4I9 A0A1B0FNR1 A0A0P6ASR2 B0W9B1 A0A1B2M4X3 A0A1A9V8U1 A0A1I8Q3N3 A0A0P5V7I4 A0A2J7QZP5 A0A0N8ACN7 A0A087ZZ66 A0A067QPZ1 D3TRK6 A0A232FFF2
A0A2W1BKB2 A0A291P0T8 A0A212F8V9 A0A2H1W3A4 K7J405 A0A0L7R1J1 D6WTS5 A0A026WFS5 A0A195CK08 A0A158NAF6 A0A195EVL4 A0A2J7Q2J5 A0A232EVU0 A0A2A3ES93 A0A087ZZR5 A0A0N0BDG0 A0A067REM7 E2A7P0 A0A151X4Z0 A0A154PIR2 E2BWS9 E0VSM5 A0A0J7KZD8 J3JZ53 U4U7N8 N6TT69 F4WMJ5 K7IP18 A0A0L7LW80 A0A232F739 A0A232EUU6 K7INV3 A0A1B6CPV8 A0A151I118 K7J407 A0A0P6FC08 A0A140CVV2 A0A336KNX9 A0A164ZMS8 A0A310S6U0 A0A182QB38 A0A2J7QCW6 E9GI56 A0A0A9YK38 D6WTU2 A0A1A9XM90 A0A182NKU4 A0A0N8DLF9 A0A182V201 A0A164KJP0 A0A182XK76 A0A0P6K0A8 A0A0P5DG11 Q7QDK4 A0A182HJR2 A0A0N8DQI7 A0A0P5DMC5 A0A182M2M9 A0A1B0BNU1 A0A164ZMP8 A0A0N8AMB7 A0A182W0L0 A0A182KWY7 A0A182R804 E2BWT3 A0A0P5BK85 B4MBK5 A0A1Y1MCE5 A0A0P4ZBC9 A0A0N8AEZ3 A0A026WFT0 A0A0P5YA29 A0A0P5M6K1 A0A0P5FT22 A0A0P5FM74 A0A0P6JPS0 A0A195CLU3 B3NYU4 A0A182IP00 A0A182Y4I9 A0A1B0FNR1 A0A0P6ASR2 B0W9B1 A0A1B2M4X3 A0A1A9V8U1 A0A1I8Q3N3 A0A0P5V7I4 A0A2J7QZP5 A0A0N8ACN7 A0A087ZZ66 A0A067QPZ1 D3TRK6 A0A232FFF2
PDB
5KET
E-value=6.31743e-84,
Score=791
Ontologies
GO
Topology
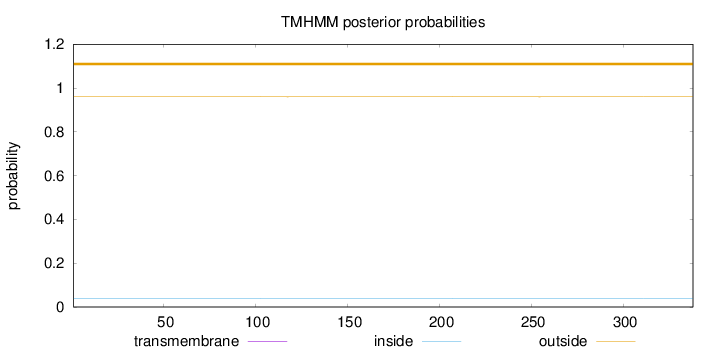
Length:
338
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0013
Exp number, first 60 AAs:
0.0005
Total prob of N-in:
0.04014
outside
1 - 338
Population Genetic Test Statistics
Pi
194.50056
Theta
171.90871
Tajima's D
0.464794
CLR
0.969387
CSRT
0.504324783760812
Interpretation
Uncertain
