Gene
KWMTBOMO15291
Pre Gene Modal
BGIBMGA005152
Annotation
PREDICTED:_N-alpha-acetyltransferase_40-like_[Plutella_xylostella]
Full name
N-alpha-acetyltransferase 40
Alternative Name
N-acetyltransferase 11
N-alpha-acetyltransferase D
N-alpha-acetyltransferase D
Location in the cell
Cytoplasmic Reliability : 1.964
Sequence
CDS
ATGGGAAAAAAATCAATAACAGCTACAAACAAACAGAAAGATAAGCGGCTTGCTAGAAAGCAAGAACAGAGGCGCATTGCTGATGCTATGACCCACGTTACAACTGCAAATAAACTTCAAGACTTGGCATCCTTGTGCAAAGAATTGTTAGTTTTCCGTAATCTCGACCTGGAGGTAGACATGTATATACAAAAGGTCACTGAACTGGATAAAAAGGTGTTACAGTGGGCCATAGACTTGACTGAAAAAAATATGAAGACACTCTATGAAACTTGTGCATGGGGTTGGAATCCTAATCGTAAGCTGGAGGAAATGACTGATGATGCAGCCTGGTATCTGATAGCCCGTGAGAAAAATGGAACACTCCTAGCTTTCTCACACTTCAGATTTGACCTGGACTTTGGAGAACCAGTATTGTATTGGTAA
Protein
MGKKSITATNKQKDKRLARKQEQRRIADAMTHVTTANKLQDLASLCKELLVFRNLDLEVDMYIQKVTELDKKVLQWAIDLTEKNMKTLYETCAWGWNPNRKLEEMTDDAAWYLIAREKNGTLLAFSHFRFDLDFGEPVLYW
Summary
Description
N-alpha-acetyltransferase that specifically mediates the acetylation of the N-terminal residues of histones H4 and H2A. In contrast to other N-alpha-acetyltransferase, has a very specific selectivity for histones H4 and H2A N-terminus and specifically recognizes the 'Ser-Gly-Arg-Gly sequence'.
Catalytic Activity
acetyl-CoA + N-terminal L-seryl-[histone H4] = CoA + H(+) + N-terminal N(alpha)-acetyl-L-seryl-[histone H4]
acetyl-CoA + N-terminal L-seryl-[histone H2A] = CoA + H(+) + N-terminal N(alpha)-acetyl-L-seryl-[histone H2A]
acetyl-CoA + N-terminal L-seryl-[histone H2A] = CoA + H(+) + N-terminal N(alpha)-acetyl-L-seryl-[histone H2A]
Similarity
Belongs to the acetyltransferase family. NAA40 subfamily.
Keywords
Acyltransferase
Complete proteome
Cytoplasm
Lipoprotein
Myristate
Nucleus
Reference proteome
Transferase
Feature
chain N-alpha-acetyltransferase 40
Uniprot
H9J6L0
A0A2H1W400
A0A2A4IXI7
A0A212F8V5
A0A194PGH2
S4PDK5
+ More
A0A0L7LVG0 A0A2W1BRS2 A0A194RKN7 A0A0T6B9J1 D6WTU5 A0A1Y1LHG7 N6TUE7 A0A2P8ZEI7 A0A1W4X8Y2 A0A1B6E3A6 A0A1S3J8M6 A0A0S7KNX7 R4V491 R4V3V0 E7EYF8 A0A3Q1D0E5 A0A3P9MUY8 A0A3B3WZN0 A0A087X6M3 A0A3B3VI12 A0A3Q2DZF4 A0A0A9ZD19 A0A0S7KNY5 A0A146Z5U4 A0A146NIG4 A0A2J7Q5I5 A0A2G9RV71 H3AVZ7 A0A146NVT9 A0A3Q2SVT0 A0A0A0A321 Q568K5 A0A3Q0QP70 H2SHB1 F6Q9B3 A0A2D4IQR2 A0A3B4GSV5 A0A3Q2WX80 A0A3P8QP87 A0A3P9B3H2 A0A3Q3ADL3 H9GIW2 A0A1A7YZE9 A0A1A8CJ69 A0A1L8GJR5 Q6NUH2 A0A1A8K9P1 A0A1A8GRI1 A0A1A8UQ09 A0A1S2ZFJ1 A0A1A8RT88 A0A1A8MR23 A0A2K5E9L4 A0A1D5P6G2 D3ZZQ5 M4ARI7 F6TZV6 A0A315VUX3 A0A2I4C3D1 Q5R9Y8 A0A067RJN6 B1H2T3 A0A3Q1FJ56 A0A3B5AZ00 A0A3P8WFH8 A0A091MH54 A0A3B4YSS0 A0A3B4VGJ7 A0A0P4WKD8 A0A3Q7R2G9 M3WL36 A0A2Y9JU26 A0A3Q7SH25 A0A2U3VH14 M3YPG1 A0A3Q7UG47 A0A224XNP9 A0A0D9R3I5 A0A1S3D588 H3D8J1 A0A2U4B4P0 A0A2Y9PNV1 A0A2I2Y7F3 A0A2Y9EIG7 Q4S0U7 A0A3Q7SWT3 I3MH75 A0A2K6C2Z0 L5KQC7 A0A2K5U6T7 A0A2K5K7P8 A0A2K6FV15 H9FZX2
A0A0L7LVG0 A0A2W1BRS2 A0A194RKN7 A0A0T6B9J1 D6WTU5 A0A1Y1LHG7 N6TUE7 A0A2P8ZEI7 A0A1W4X8Y2 A0A1B6E3A6 A0A1S3J8M6 A0A0S7KNX7 R4V491 R4V3V0 E7EYF8 A0A3Q1D0E5 A0A3P9MUY8 A0A3B3WZN0 A0A087X6M3 A0A3B3VI12 A0A3Q2DZF4 A0A0A9ZD19 A0A0S7KNY5 A0A146Z5U4 A0A146NIG4 A0A2J7Q5I5 A0A2G9RV71 H3AVZ7 A0A146NVT9 A0A3Q2SVT0 A0A0A0A321 Q568K5 A0A3Q0QP70 H2SHB1 F6Q9B3 A0A2D4IQR2 A0A3B4GSV5 A0A3Q2WX80 A0A3P8QP87 A0A3P9B3H2 A0A3Q3ADL3 H9GIW2 A0A1A7YZE9 A0A1A8CJ69 A0A1L8GJR5 Q6NUH2 A0A1A8K9P1 A0A1A8GRI1 A0A1A8UQ09 A0A1S2ZFJ1 A0A1A8RT88 A0A1A8MR23 A0A2K5E9L4 A0A1D5P6G2 D3ZZQ5 M4ARI7 F6TZV6 A0A315VUX3 A0A2I4C3D1 Q5R9Y8 A0A067RJN6 B1H2T3 A0A3Q1FJ56 A0A3B5AZ00 A0A3P8WFH8 A0A091MH54 A0A3B4YSS0 A0A3B4VGJ7 A0A0P4WKD8 A0A3Q7R2G9 M3WL36 A0A2Y9JU26 A0A3Q7SH25 A0A2U3VH14 M3YPG1 A0A3Q7UG47 A0A224XNP9 A0A0D9R3I5 A0A1S3D588 H3D8J1 A0A2U4B4P0 A0A2Y9PNV1 A0A2I2Y7F3 A0A2Y9EIG7 Q4S0U7 A0A3Q7SWT3 I3MH75 A0A2K6C2Z0 L5KQC7 A0A2K5U6T7 A0A2K5K7P8 A0A2K6FV15 H9FZX2
EC Number
2.3.1.257
Pubmed
19121390
22118469
26354079
23622113
26227816
28756777
+ More
18362917 19820115 28004739 23537049 29403074 23594743 25401762 26823975 9215903 21551351 17495919 25186727 21881562 27762356 15592404 15057822 15632090 23542700 25243066 29703783 24845553 20431018 24487278 17975172 15496914 22398555 23258410 25319552
18362917 19820115 28004739 23537049 29403074 23594743 25401762 26823975 9215903 21551351 17495919 25186727 21881562 27762356 15592404 15057822 15632090 23542700 25243066 29703783 24845553 20431018 24487278 17975172 15496914 22398555 23258410 25319552
EMBL
BABH01019867
ODYU01006049
SOQ47562.1
NWSH01005189
PCG64369.1
AGBW02009658
+ More
OWR50185.1 KQ459604 KPI92402.1 GAIX01003641 JAA88919.1 JTDY01000014 KOB79420.1 KZ149994 PZC75480.1 KQ460045 KPJ18102.1 LJIG01003015 KRT83925.1 KQ971352 EFA07325.1 GEZM01055207 JAV73089.1 APGK01018323 APGK01018324 APGK01018325 KB740073 KB632222 ENN81673.1 ERL90203.1 PYGN01000079 PSN54922.1 GEDC01004894 JAS32404.1 GBYX01202420 GBYX01202419 JAO79047.1 KC740844 AGM32668.1 KC571989 AGM32488.1 CABZ01043953 AYCK01016166 GBHO01002366 GBRD01018121 GDHC01002865 JAG41238.1 JAG47706.1 JAQ15764.1 GBYX01202423 GBYX01202422 JAO79045.1 GCES01025256 JAR61067.1 GCES01155067 JAQ31255.1 NEVH01017557 PNF23835.1 KV929863 PIO31113.1 AFYH01015748 GCES01150448 JAQ35874.1 KL870439 KGL88446.1 BC092819 IACK01118948 LAA86591.1 AAWZ02032030 HADW01001147 HADX01013264 SBP35496.1 HADZ01014865 HAEA01005438 SBP78806.1 CM004472 OCT84087.1 BC068615 HAED01002534 HAEE01008871 SBR28921.1 HAEB01021349 HAEC01005690 SBQ73767.1 HADY01013365 HAEJ01008960 SBS49417.1 HAEI01009692 SBS08484.1 HAEF01017862 HAEG01015413 SBR59021.1 AADN05000754 AC126148 CH473953 EDM12671.1 GAMT01001998 GAMS01005015 GAMR01009740 GAMQ01005392 GAMP01010297 JAB09863.1 JAB18121.1 JAB24192.1 JAB36459.1 JAB42458.1 NHOQ01001086 PWA27160.1 CR859242 CAH91422.1 KK852428 KDR24066.1 AAMC01118463 BC161120 AAI61120.1 KK504630 KFP60915.1 GDRN01083168 JAI61709.1 AANG04000779 AEYP01074286 AEYP01074287 AEYP01074288 GFTR01003712 JAW12714.1 AQIB01078872 AQIB01078873 AQIB01078874 AQIB01078875 CABD030079646 CABD030079647 CABD030079648 CABD030079649 CAAE01014779 CAG05735.1 AGTP01072483 AGTP01072484 KB030625 ELK13410.1 AQIA01019940 AQIA01019941 AQIA01019942 JU336428 JU477242 JU477243 JU477244 JU477245 JV048279 AFE80181.1 AFH34046.1 AFI38350.1
OWR50185.1 KQ459604 KPI92402.1 GAIX01003641 JAA88919.1 JTDY01000014 KOB79420.1 KZ149994 PZC75480.1 KQ460045 KPJ18102.1 LJIG01003015 KRT83925.1 KQ971352 EFA07325.1 GEZM01055207 JAV73089.1 APGK01018323 APGK01018324 APGK01018325 KB740073 KB632222 ENN81673.1 ERL90203.1 PYGN01000079 PSN54922.1 GEDC01004894 JAS32404.1 GBYX01202420 GBYX01202419 JAO79047.1 KC740844 AGM32668.1 KC571989 AGM32488.1 CABZ01043953 AYCK01016166 GBHO01002366 GBRD01018121 GDHC01002865 JAG41238.1 JAG47706.1 JAQ15764.1 GBYX01202423 GBYX01202422 JAO79045.1 GCES01025256 JAR61067.1 GCES01155067 JAQ31255.1 NEVH01017557 PNF23835.1 KV929863 PIO31113.1 AFYH01015748 GCES01150448 JAQ35874.1 KL870439 KGL88446.1 BC092819 IACK01118948 LAA86591.1 AAWZ02032030 HADW01001147 HADX01013264 SBP35496.1 HADZ01014865 HAEA01005438 SBP78806.1 CM004472 OCT84087.1 BC068615 HAED01002534 HAEE01008871 SBR28921.1 HAEB01021349 HAEC01005690 SBQ73767.1 HADY01013365 HAEJ01008960 SBS49417.1 HAEI01009692 SBS08484.1 HAEF01017862 HAEG01015413 SBR59021.1 AADN05000754 AC126148 CH473953 EDM12671.1 GAMT01001998 GAMS01005015 GAMR01009740 GAMQ01005392 GAMP01010297 JAB09863.1 JAB18121.1 JAB24192.1 JAB36459.1 JAB42458.1 NHOQ01001086 PWA27160.1 CR859242 CAH91422.1 KK852428 KDR24066.1 AAMC01118463 BC161120 AAI61120.1 KK504630 KFP60915.1 GDRN01083168 JAI61709.1 AANG04000779 AEYP01074286 AEYP01074287 AEYP01074288 GFTR01003712 JAW12714.1 AQIB01078872 AQIB01078873 AQIB01078874 AQIB01078875 CABD030079646 CABD030079647 CABD030079648 CABD030079649 CAAE01014779 CAG05735.1 AGTP01072483 AGTP01072484 KB030625 ELK13410.1 AQIA01019940 AQIA01019941 AQIA01019942 JU336428 JU477242 JU477243 JU477244 JU477245 JV048279 AFE80181.1 AFH34046.1 AFI38350.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000037510
UP000053240
+ More
UP000007266 UP000019118 UP000030742 UP000245037 UP000192223 UP000085678 UP000000437 UP000257160 UP000242638 UP000261480 UP000028760 UP000261500 UP000265020 UP000265000 UP000235965 UP000008672 UP000053858 UP000261340 UP000005226 UP000002280 UP000261460 UP000264840 UP000265100 UP000265160 UP000264800 UP000001646 UP000186698 UP000079721 UP000233020 UP000000539 UP000002494 UP000002852 UP000008225 UP000192220 UP000027135 UP000008143 UP000257200 UP000261400 UP000265120 UP000261360 UP000261420 UP000286641 UP000011712 UP000248482 UP000286640 UP000245340 UP000000715 UP000286642 UP000029965 UP000079169 UP000007303 UP000245320 UP000248483 UP000001519 UP000248484 UP000005215 UP000233120 UP000010552 UP000233100 UP000233080 UP000233160
UP000007266 UP000019118 UP000030742 UP000245037 UP000192223 UP000085678 UP000000437 UP000257160 UP000242638 UP000261480 UP000028760 UP000261500 UP000265020 UP000265000 UP000235965 UP000008672 UP000053858 UP000261340 UP000005226 UP000002280 UP000261460 UP000264840 UP000265100 UP000265160 UP000264800 UP000001646 UP000186698 UP000079721 UP000233020 UP000000539 UP000002494 UP000002852 UP000008225 UP000192220 UP000027135 UP000008143 UP000257200 UP000261400 UP000265120 UP000261360 UP000261420 UP000286641 UP000011712 UP000248482 UP000286640 UP000245340 UP000000715 UP000286642 UP000029965 UP000079169 UP000007303 UP000245320 UP000248483 UP000001519 UP000248484 UP000005215 UP000233120 UP000010552 UP000233100 UP000233080 UP000233160
Pfam
PF00583 Acetyltransf_1
SUPFAM
SSF55729
SSF55729
ProteinModelPortal
H9J6L0
A0A2H1W400
A0A2A4IXI7
A0A212F8V5
A0A194PGH2
S4PDK5
+ More
A0A0L7LVG0 A0A2W1BRS2 A0A194RKN7 A0A0T6B9J1 D6WTU5 A0A1Y1LHG7 N6TUE7 A0A2P8ZEI7 A0A1W4X8Y2 A0A1B6E3A6 A0A1S3J8M6 A0A0S7KNX7 R4V491 R4V3V0 E7EYF8 A0A3Q1D0E5 A0A3P9MUY8 A0A3B3WZN0 A0A087X6M3 A0A3B3VI12 A0A3Q2DZF4 A0A0A9ZD19 A0A0S7KNY5 A0A146Z5U4 A0A146NIG4 A0A2J7Q5I5 A0A2G9RV71 H3AVZ7 A0A146NVT9 A0A3Q2SVT0 A0A0A0A321 Q568K5 A0A3Q0QP70 H2SHB1 F6Q9B3 A0A2D4IQR2 A0A3B4GSV5 A0A3Q2WX80 A0A3P8QP87 A0A3P9B3H2 A0A3Q3ADL3 H9GIW2 A0A1A7YZE9 A0A1A8CJ69 A0A1L8GJR5 Q6NUH2 A0A1A8K9P1 A0A1A8GRI1 A0A1A8UQ09 A0A1S2ZFJ1 A0A1A8RT88 A0A1A8MR23 A0A2K5E9L4 A0A1D5P6G2 D3ZZQ5 M4ARI7 F6TZV6 A0A315VUX3 A0A2I4C3D1 Q5R9Y8 A0A067RJN6 B1H2T3 A0A3Q1FJ56 A0A3B5AZ00 A0A3P8WFH8 A0A091MH54 A0A3B4YSS0 A0A3B4VGJ7 A0A0P4WKD8 A0A3Q7R2G9 M3WL36 A0A2Y9JU26 A0A3Q7SH25 A0A2U3VH14 M3YPG1 A0A3Q7UG47 A0A224XNP9 A0A0D9R3I5 A0A1S3D588 H3D8J1 A0A2U4B4P0 A0A2Y9PNV1 A0A2I2Y7F3 A0A2Y9EIG7 Q4S0U7 A0A3Q7SWT3 I3MH75 A0A2K6C2Z0 L5KQC7 A0A2K5U6T7 A0A2K5K7P8 A0A2K6FV15 H9FZX2
A0A0L7LVG0 A0A2W1BRS2 A0A194RKN7 A0A0T6B9J1 D6WTU5 A0A1Y1LHG7 N6TUE7 A0A2P8ZEI7 A0A1W4X8Y2 A0A1B6E3A6 A0A1S3J8M6 A0A0S7KNX7 R4V491 R4V3V0 E7EYF8 A0A3Q1D0E5 A0A3P9MUY8 A0A3B3WZN0 A0A087X6M3 A0A3B3VI12 A0A3Q2DZF4 A0A0A9ZD19 A0A0S7KNY5 A0A146Z5U4 A0A146NIG4 A0A2J7Q5I5 A0A2G9RV71 H3AVZ7 A0A146NVT9 A0A3Q2SVT0 A0A0A0A321 Q568K5 A0A3Q0QP70 H2SHB1 F6Q9B3 A0A2D4IQR2 A0A3B4GSV5 A0A3Q2WX80 A0A3P8QP87 A0A3P9B3H2 A0A3Q3ADL3 H9GIW2 A0A1A7YZE9 A0A1A8CJ69 A0A1L8GJR5 Q6NUH2 A0A1A8K9P1 A0A1A8GRI1 A0A1A8UQ09 A0A1S2ZFJ1 A0A1A8RT88 A0A1A8MR23 A0A2K5E9L4 A0A1D5P6G2 D3ZZQ5 M4ARI7 F6TZV6 A0A315VUX3 A0A2I4C3D1 Q5R9Y8 A0A067RJN6 B1H2T3 A0A3Q1FJ56 A0A3B5AZ00 A0A3P8WFH8 A0A091MH54 A0A3B4YSS0 A0A3B4VGJ7 A0A0P4WKD8 A0A3Q7R2G9 M3WL36 A0A2Y9JU26 A0A3Q7SH25 A0A2U3VH14 M3YPG1 A0A3Q7UG47 A0A224XNP9 A0A0D9R3I5 A0A1S3D588 H3D8J1 A0A2U4B4P0 A0A2Y9PNV1 A0A2I2Y7F3 A0A2Y9EIG7 Q4S0U7 A0A3Q7SWT3 I3MH75 A0A2K6C2Z0 L5KQC7 A0A2K5U6T7 A0A2K5K7P8 A0A2K6FV15 H9FZX2
PDB
4U9W
E-value=5.99226e-21,
Score=242
Ontologies
KEGG
GO
PANTHER
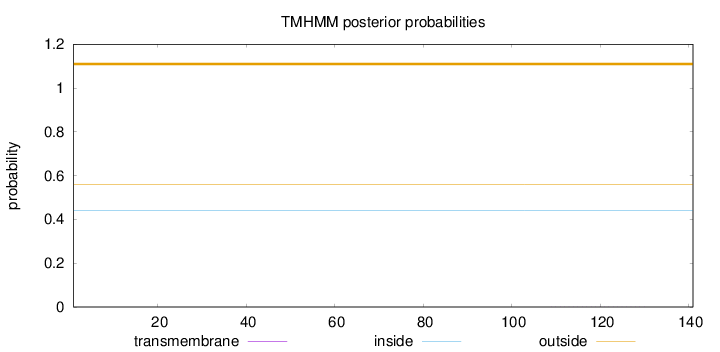
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
Length:
141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00706
Exp number, first 60 AAs:
0.00012
Total prob of N-in:
0.44189
outside
1 - 141
Population Genetic Test Statistics
Pi
260.378495
Theta
187.37093
Tajima's D
1.074208
CLR
0.016194
CSRT
0.67691615419229
Interpretation
Uncertain
