Gene
KWMTBOMO15290
Pre Gene Modal
BGIBMGA005046
Annotation
UDP-glycosyltransferase_UGT47A1_precursor_[Bombyx_mori]
Full name
UDP-glucuronosyltransferase
Location in the cell
PlasmaMembrane Reliability : 2.522
Sequence
CDS
ATGCGGGTGGCGTGGCTGCTGTGGTTAGCGACGGTCGCACGCGCTGCTCGTCTGCTCGTGGTGCTGCCCACAAACACTAAGAGCCATTACGCCATGTATAGCAGGCTTATTGAGGCGCTAGCGAAAAGGGACCATCAACTTACCGTTATCACACACTTCCCTGTGGACGCACCGCAACCTAACATTAATCAAATAAGCTTAGCTGGAACAATACCGGAGATCACTAATAATCTTACAAGAAAATATGATTCTTTGAAGCCGAACTTCATCCGCAATTTGGAACAAATCATCAGTGAATGCGTTAATGCTTGCGAAACAGTAGTACAGCAAGATTCCTTCAAAGAGTTACTTAATTCGACCGCTTCTTTTGATTTAATTATCGTAGAAGTCTTCGGAAGTGATTGTTTTTTACCTTTAGGTCATAGATTCCGGGCCCCCGTTGTCGGATTATTGTCTAGCGTCCCGCTACCTTGGGTCAATGATTACCTTGGCAACCCAGAAACAGCGTCTTACATACCGTCCTACATGATGGGTTATGGACAACACATGAGCCTATGGGAACGATTATCAAACACAATAGCTATTATATTAGCTAAGATTTTATACACATACAAATCAAGGATTCCTTCACAAGTGATAGTAGACAGAGTCTTCGGACATGGAAATAACTTACAAAAGCTCGCGAAAAACTACAGTGTGATTTTATCTAACAGTCACTTTAGCATTAACGAAGTCCGACCTCTTGTGCCTGGTCTAGTAGAAGTAGGCGGTCTACATTTGGATCACAGTCAAACACTTCCTAAGAATATGAAGAAACTTTTAGATGCGTCAACAGACGGTGTCATCTACTGGAGCTTTGGCTCGATGTCCAGAATAGAAACGATACCTAGCGAAAAGCTTTCAGGTATATTTGAAGCAATATCTGAACTACCACAATTGGTCTTCGTGAAGATGGATAGAAGAAGGCTCACAAAGAATATCACTGTGCCTGACAATGTATATACTATGGATTGGATTCCGCAGTATGCTACATTATGTCACCCAAACGTCAAGTTATTCATTTCTCATGGAGGCTTGCTTGGCACACAGGAAGCAGTGGCTTGTGGGGTACCTATGTTAATGGTGCCTCTATATGCTGACCAAGCTCTTAATTCTCAGTCTATGTTTGATAAAAACGTTGCCAGAATTCTGGATCTACACGAGGCTGATAAATATGAATGGAAGAGAGCTTTACATGATTTATTGAGTAACAAAAAGTATAGAGAAAATTCAAAAACGCTAAAAGAAATATTTTTAGATAGACCAATAAATCCTTTAGATATGGGCGTCTACTGGATTGAATATGTGCTGAGATACCGAGATATTAAAAATAAATAA
Protein
MRVAWLLWLATVARAARLLVVLPTNTKSHYAMYSRLIEALAKRDHQLTVITHFPVDAPQPNINQISLAGTIPEITNNLTRKYDSLKPNFIRNLEQIISECVNACETVVQQDSFKELLNSTASFDLIIVEVFGSDCFLPLGHRFRAPVVGLLSSVPLPWVNDYLGNPETASYIPSYMMGYGQHMSLWERLSNTIAIILAKILYTYKSRIPSQVIVDRVFGHGNNLQKLAKNYSVILSNSHFSINEVRPLVPGLVEVGGLHLDHSQTLPKNMKKLLDASTDGVIYWSFGSMSRIETIPSEKLSGIFEAISELPQLVFVKMDRRRLTKNITVPDNVYTMDWIPQYATLCHPNVKLFISHGGLLGTQEAVACGVPMLMVPLYADQALNSQSMFDKNVARILDLHEADKYEWKRALHDLLSNKKYRENSKTLKEIFLDRPINPLDMGVYWIEYVLRYRDIKNK
Summary
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Belongs to the Ca(2+):cation antiporter (CaCA) (TC 2.A.19) family.
Belongs to the Ca(2+):cation antiporter (CaCA) (TC 2.A.19) family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
G9LPW3
G9LPR9
A0A2W1BMF5
A0A2A4IYW0
A0A2H1W3F1
A0A286MXP7
+ More
A0A194PG87 A0A1L8D6T1 A0A194RJU8 A0A212F8W5 A0A2J7QFA7 A0A067RIR8 A0A139WIV0 D2A2K6 A0A2P8ZG63 A0A2J7R4S8 A0A1W4XRE9 A0A2J7QFA8 J3JYV9 A0A067RAD1 A0A1W4W691 A0A1W4W6Y1 A0A2J7R4W8 A0A139WIV3 A0A139WIR0 A0A0T6BCY2 A0A2P8YRX1 A0A2J7R4V4 V5H2B2 A0A1W4WNP2 A0A2J7R4T0 A0A2J7R4V3 A0A0L7QTK6 A0A2J7R4V1 A0A1B6F5J1 A0A087ZQF4 D2A2V5 A0A2P8ZG78 A0A2J7R4W4 A0A1B6EZ83 A0A2A3EGP2 A0A2P8ZG70 A0A1B6KW89 A0A0C9RNU1 A0A1W4X720 A0A1B6CX28 A0A1B6LEK5 E2C3N6 K7JB17 A0A067RFX2 A0A139WIW7 A0A151WM71 F4WPC2 A0A067RAP7 A0A067QGQ4 A0A2J7R4W2 A0A1B6K0J6 A0A1B6K3R7 A0A1B6FAC1 A0A2P8ZG71 A0A232EQT2 A0A195E5I6 A0A1B6KV12 A0A158NQX4 A0A1B6LAB9 A0A3L8DPI9 A0A026WNX0 A0A1B6I336 A0A3L8DXJ2 A0A151WXP6 A0A1B6EUC3 A0A1B6FW51 A0A2J7Q396 A0A195B191 A0A026WYL1 A0A2S2NJQ5 A0A0L7QNV9 A0A1W4WYE4 E0VGC0 A0A2S2NGA3 A0A1B6KXR8 E2BN43 A0A2H8TPC9 A0A2D1GSK1 A0A151ICV8 J9JN70 A0A182F4M1 A0A1B6JYM7 W5J9T2 A0A2D1GSK5 A0A3L8DYP6 A0A1B6ET74 A0A026WMH2 A0A026WPZ7 A0A1B6IMF2 A0A1B6IK98 A0A0L7QNF8 A0A1W4WMC9 E9IV89
A0A194PG87 A0A1L8D6T1 A0A194RJU8 A0A212F8W5 A0A2J7QFA7 A0A067RIR8 A0A139WIV0 D2A2K6 A0A2P8ZG63 A0A2J7R4S8 A0A1W4XRE9 A0A2J7QFA8 J3JYV9 A0A067RAD1 A0A1W4W691 A0A1W4W6Y1 A0A2J7R4W8 A0A139WIV3 A0A139WIR0 A0A0T6BCY2 A0A2P8YRX1 A0A2J7R4V4 V5H2B2 A0A1W4WNP2 A0A2J7R4T0 A0A2J7R4V3 A0A0L7QTK6 A0A2J7R4V1 A0A1B6F5J1 A0A087ZQF4 D2A2V5 A0A2P8ZG78 A0A2J7R4W4 A0A1B6EZ83 A0A2A3EGP2 A0A2P8ZG70 A0A1B6KW89 A0A0C9RNU1 A0A1W4X720 A0A1B6CX28 A0A1B6LEK5 E2C3N6 K7JB17 A0A067RFX2 A0A139WIW7 A0A151WM71 F4WPC2 A0A067RAP7 A0A067QGQ4 A0A2J7R4W2 A0A1B6K0J6 A0A1B6K3R7 A0A1B6FAC1 A0A2P8ZG71 A0A232EQT2 A0A195E5I6 A0A1B6KV12 A0A158NQX4 A0A1B6LAB9 A0A3L8DPI9 A0A026WNX0 A0A1B6I336 A0A3L8DXJ2 A0A151WXP6 A0A1B6EUC3 A0A1B6FW51 A0A2J7Q396 A0A195B191 A0A026WYL1 A0A2S2NJQ5 A0A0L7QNV9 A0A1W4WYE4 E0VGC0 A0A2S2NGA3 A0A1B6KXR8 E2BN43 A0A2H8TPC9 A0A2D1GSK1 A0A151ICV8 J9JN70 A0A182F4M1 A0A1B6JYM7 W5J9T2 A0A2D1GSK5 A0A3L8DYP6 A0A1B6ET74 A0A026WMH2 A0A026WPZ7 A0A1B6IMF2 A0A1B6IK98 A0A0L7QNF8 A0A1W4WMC9 E9IV89
EC Number
2.4.1.17
Pubmed
EMBL
BABH01019867
JQ070270
AEW43186.1
JQ070226
AEW43142.1
KZ149994
+ More
PZC75471.1 NWSH01005189 PCG64370.1 ODYU01006049 SOQ47563.1 MF684365 ASX93999.1 KQ459604 KPI92401.1 KY202926 GEYN01000075 AUC64270.1 JAV02054.1 KQ460045 KPJ18103.1 AGBW02009658 OWR50184.1 NEVH01015300 PNF27275.1 KK852607 KDR20346.1 KQ971338 KYB27844.1 EFA02010.1 PYGN01000068 PSN55493.1 NEVH01007402 PNF35836.1 PNF27276.1 BT128440 AEE63397.1 KK852590 KDR20744.1 PNF35870.1 KYB27842.1 KYB27843.1 LJIG01001838 KRT85102.1 PYGN01000401 PSN46987.1 PNF35866.1 GALX01001513 JAB66953.1 PNF35837.1 PNF35864.1 KQ414747 KOC61874.1 PNF35863.1 GECZ01024322 GECZ01008492 JAS45447.1 JAS61277.1 EFA02977.2 PSN55501.1 PNF35869.1 GECZ01026545 JAS43224.1 KZ288262 PBC30389.1 PSN55494.1 GEBQ01024251 JAT15726.1 GBYB01008756 JAG78523.1 GEDC01019423 GEDC01014974 JAS17875.1 JAS22324.1 GEBQ01017854 JAT22123.1 GL452328 EFN77459.1 AAZX01012567 KK852488 KDR22771.1 KYB27856.1 KQ982944 KYQ48964.1 GL888243 EGI64017.1 KDR20745.1 KK853490 KDR07235.1 PNF35872.1 GECU01002730 JAT04977.1 GECU01001605 JAT06102.1 GECZ01022678 JAS47091.1 PSN55491.1 NNAY01002740 OXU20656.1 KQ979608 KYN20423.1 GEBQ01024697 JAT15280.1 ADTU01023695 GEBQ01019317 JAT20660.1 QOIP01000005 RLU22281.1 KK107139 EZA57757.1 GECU01026373 JAS81333.1 QOIP01000003 RLU24953.1 KQ982668 KYQ52547.1 GECZ01028213 JAS41556.1 GECZ01015337 JAS54432.1 NEVH01019069 PNF23054.1 KQ976691 KYM77969.1 KK107063 EZA61120.1 GGMR01004776 MBY17395.1 KQ414851 KOC60176.1 DS235135 EEB12426.1 GGMR01003614 MBY16233.1 GEBQ01023933 JAT16044.1 GL449382 EFN82947.1 GFXV01004222 MBW16027.1 MF471423 ATN96029.1 KQ978002 KYM98244.1 ABLF02040513 GECU01003693 JAT04014.1 ADMH02001962 ETN60168.1 MF471449 ATN96055.1 RLU24928.1 GECZ01028564 JAS41205.1 KK107151 EZA57272.1 EZA57756.1 RLU24627.1 GECU01019610 JAS88096.1 GECU01020359 JAS87347.1 KOC60177.1 GL766176 EFZ15506.1
PZC75471.1 NWSH01005189 PCG64370.1 ODYU01006049 SOQ47563.1 MF684365 ASX93999.1 KQ459604 KPI92401.1 KY202926 GEYN01000075 AUC64270.1 JAV02054.1 KQ460045 KPJ18103.1 AGBW02009658 OWR50184.1 NEVH01015300 PNF27275.1 KK852607 KDR20346.1 KQ971338 KYB27844.1 EFA02010.1 PYGN01000068 PSN55493.1 NEVH01007402 PNF35836.1 PNF27276.1 BT128440 AEE63397.1 KK852590 KDR20744.1 PNF35870.1 KYB27842.1 KYB27843.1 LJIG01001838 KRT85102.1 PYGN01000401 PSN46987.1 PNF35866.1 GALX01001513 JAB66953.1 PNF35837.1 PNF35864.1 KQ414747 KOC61874.1 PNF35863.1 GECZ01024322 GECZ01008492 JAS45447.1 JAS61277.1 EFA02977.2 PSN55501.1 PNF35869.1 GECZ01026545 JAS43224.1 KZ288262 PBC30389.1 PSN55494.1 GEBQ01024251 JAT15726.1 GBYB01008756 JAG78523.1 GEDC01019423 GEDC01014974 JAS17875.1 JAS22324.1 GEBQ01017854 JAT22123.1 GL452328 EFN77459.1 AAZX01012567 KK852488 KDR22771.1 KYB27856.1 KQ982944 KYQ48964.1 GL888243 EGI64017.1 KDR20745.1 KK853490 KDR07235.1 PNF35872.1 GECU01002730 JAT04977.1 GECU01001605 JAT06102.1 GECZ01022678 JAS47091.1 PSN55491.1 NNAY01002740 OXU20656.1 KQ979608 KYN20423.1 GEBQ01024697 JAT15280.1 ADTU01023695 GEBQ01019317 JAT20660.1 QOIP01000005 RLU22281.1 KK107139 EZA57757.1 GECU01026373 JAS81333.1 QOIP01000003 RLU24953.1 KQ982668 KYQ52547.1 GECZ01028213 JAS41556.1 GECZ01015337 JAS54432.1 NEVH01019069 PNF23054.1 KQ976691 KYM77969.1 KK107063 EZA61120.1 GGMR01004776 MBY17395.1 KQ414851 KOC60176.1 DS235135 EEB12426.1 GGMR01003614 MBY16233.1 GEBQ01023933 JAT16044.1 GL449382 EFN82947.1 GFXV01004222 MBW16027.1 MF471423 ATN96029.1 KQ978002 KYM98244.1 ABLF02040513 GECU01003693 JAT04014.1 ADMH02001962 ETN60168.1 MF471449 ATN96055.1 RLU24928.1 GECZ01028564 JAS41205.1 KK107151 EZA57272.1 EZA57756.1 RLU24627.1 GECU01019610 JAS88096.1 GECU01020359 JAS87347.1 KOC60177.1 GL766176 EFZ15506.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000235965
+ More
UP000027135 UP000007266 UP000245037 UP000192223 UP000053825 UP000005203 UP000242457 UP000008237 UP000002358 UP000075809 UP000007755 UP000215335 UP000078492 UP000005205 UP000279307 UP000053097 UP000078540 UP000009046 UP000078542 UP000007819 UP000069272 UP000000673
UP000027135 UP000007266 UP000245037 UP000192223 UP000053825 UP000005203 UP000242457 UP000008237 UP000002358 UP000075809 UP000007755 UP000215335 UP000078492 UP000005205 UP000279307 UP000053097 UP000078540 UP000009046 UP000078542 UP000007819 UP000069272 UP000000673
PRIDE
Interpro
ProteinModelPortal
G9LPW3
G9LPR9
A0A2W1BMF5
A0A2A4IYW0
A0A2H1W3F1
A0A286MXP7
+ More
A0A194PG87 A0A1L8D6T1 A0A194RJU8 A0A212F8W5 A0A2J7QFA7 A0A067RIR8 A0A139WIV0 D2A2K6 A0A2P8ZG63 A0A2J7R4S8 A0A1W4XRE9 A0A2J7QFA8 J3JYV9 A0A067RAD1 A0A1W4W691 A0A1W4W6Y1 A0A2J7R4W8 A0A139WIV3 A0A139WIR0 A0A0T6BCY2 A0A2P8YRX1 A0A2J7R4V4 V5H2B2 A0A1W4WNP2 A0A2J7R4T0 A0A2J7R4V3 A0A0L7QTK6 A0A2J7R4V1 A0A1B6F5J1 A0A087ZQF4 D2A2V5 A0A2P8ZG78 A0A2J7R4W4 A0A1B6EZ83 A0A2A3EGP2 A0A2P8ZG70 A0A1B6KW89 A0A0C9RNU1 A0A1W4X720 A0A1B6CX28 A0A1B6LEK5 E2C3N6 K7JB17 A0A067RFX2 A0A139WIW7 A0A151WM71 F4WPC2 A0A067RAP7 A0A067QGQ4 A0A2J7R4W2 A0A1B6K0J6 A0A1B6K3R7 A0A1B6FAC1 A0A2P8ZG71 A0A232EQT2 A0A195E5I6 A0A1B6KV12 A0A158NQX4 A0A1B6LAB9 A0A3L8DPI9 A0A026WNX0 A0A1B6I336 A0A3L8DXJ2 A0A151WXP6 A0A1B6EUC3 A0A1B6FW51 A0A2J7Q396 A0A195B191 A0A026WYL1 A0A2S2NJQ5 A0A0L7QNV9 A0A1W4WYE4 E0VGC0 A0A2S2NGA3 A0A1B6KXR8 E2BN43 A0A2H8TPC9 A0A2D1GSK1 A0A151ICV8 J9JN70 A0A182F4M1 A0A1B6JYM7 W5J9T2 A0A2D1GSK5 A0A3L8DYP6 A0A1B6ET74 A0A026WMH2 A0A026WPZ7 A0A1B6IMF2 A0A1B6IK98 A0A0L7QNF8 A0A1W4WMC9 E9IV89
A0A194PG87 A0A1L8D6T1 A0A194RJU8 A0A212F8W5 A0A2J7QFA7 A0A067RIR8 A0A139WIV0 D2A2K6 A0A2P8ZG63 A0A2J7R4S8 A0A1W4XRE9 A0A2J7QFA8 J3JYV9 A0A067RAD1 A0A1W4W691 A0A1W4W6Y1 A0A2J7R4W8 A0A139WIV3 A0A139WIR0 A0A0T6BCY2 A0A2P8YRX1 A0A2J7R4V4 V5H2B2 A0A1W4WNP2 A0A2J7R4T0 A0A2J7R4V3 A0A0L7QTK6 A0A2J7R4V1 A0A1B6F5J1 A0A087ZQF4 D2A2V5 A0A2P8ZG78 A0A2J7R4W4 A0A1B6EZ83 A0A2A3EGP2 A0A2P8ZG70 A0A1B6KW89 A0A0C9RNU1 A0A1W4X720 A0A1B6CX28 A0A1B6LEK5 E2C3N6 K7JB17 A0A067RFX2 A0A139WIW7 A0A151WM71 F4WPC2 A0A067RAP7 A0A067QGQ4 A0A2J7R4W2 A0A1B6K0J6 A0A1B6K3R7 A0A1B6FAC1 A0A2P8ZG71 A0A232EQT2 A0A195E5I6 A0A1B6KV12 A0A158NQX4 A0A1B6LAB9 A0A3L8DPI9 A0A026WNX0 A0A1B6I336 A0A3L8DXJ2 A0A151WXP6 A0A1B6EUC3 A0A1B6FW51 A0A2J7Q396 A0A195B191 A0A026WYL1 A0A2S2NJQ5 A0A0L7QNV9 A0A1W4WYE4 E0VGC0 A0A2S2NGA3 A0A1B6KXR8 E2BN43 A0A2H8TPC9 A0A2D1GSK1 A0A151ICV8 J9JN70 A0A182F4M1 A0A1B6JYM7 W5J9T2 A0A2D1GSK5 A0A3L8DYP6 A0A1B6ET74 A0A026WMH2 A0A026WPZ7 A0A1B6IMF2 A0A1B6IK98 A0A0L7QNF8 A0A1W4WMC9 E9IV89
PDB
2O6L
E-value=5.53725e-18,
Score=224
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
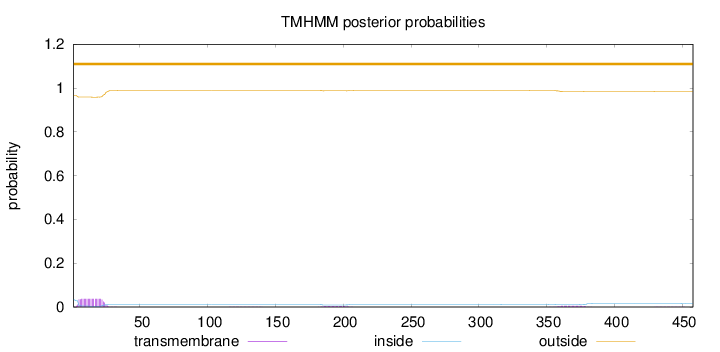
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 15,
Likelihood: 0.928762
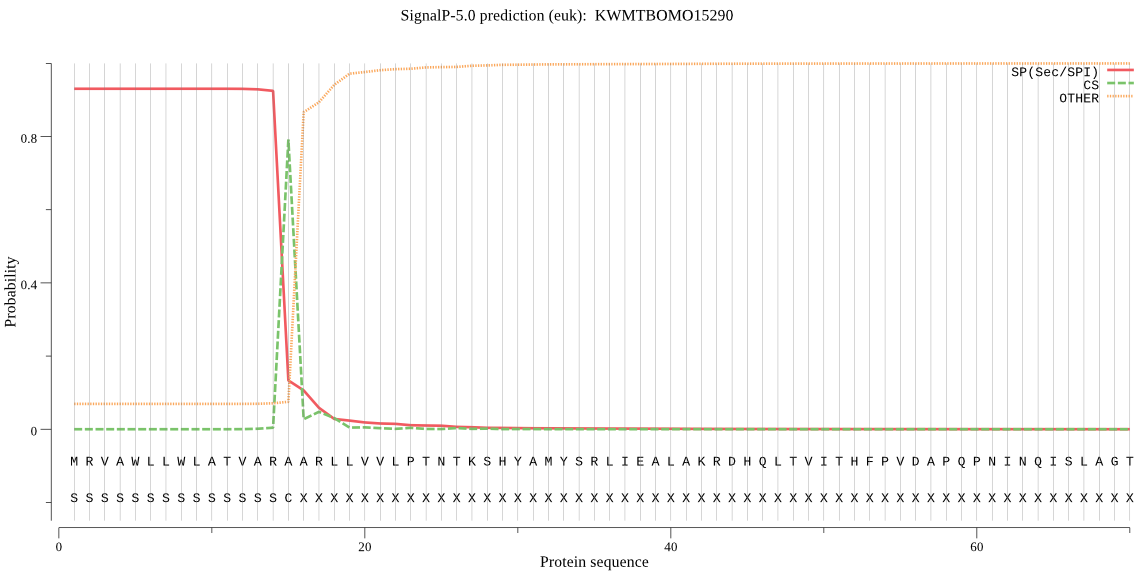
Length:
458
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.956679999999997
Exp number, first 60 AAs:
0.75059
Total prob of N-in:
0.03387
outside
1 - 458
Population Genetic Test Statistics
Pi
250.764053
Theta
185.680697
Tajima's D
0.876538
CLR
0.175301
CSRT
0.619069046547673
Interpretation
Uncertain
