Gene
KWMTBOMO15288 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005155
Annotation
alcohol_dehydrogenase_[Ostrinia_furnacalis]
Location in the cell
Cytoplasmic Reliability : 3.394
Sequence
CDS
ATGTCGGTCCCAGAATACTTCGAGTTAAACGGAGGAGATCGCATGCCCAAAATTGGATTTGGTACTTGGCAGTCATCTGATGAAGTCTTAGAGAAGGCTGTACTAGCCGCTTTGGATGCGGGCTATAGACACTTCGACACCGCCCATGCCTATGAGAACGAGGCAGCTTTGGGGCGAGCGCTACGGAAATGGATCGGTGATAACCCCGCCAAAAGGCGAGAGCTATTCGTCGTCACCAAATTGCCACCAGGAGGTAATCGTCCAGAATTGGTCAGGGAGTATTTTGACACGTCTTTCCGTAACCTCGGATTGGATTACATAGATATGTATTTGATGCACACACCCTTTGCGTTCGAACATGTACCAGGTGACCTGCATCCTAAGAACCCTGACGGCACTATGAAGGTGGACTATTCCACGGATCTAGCAGCAGTCTGGAAAACACTCCTAAAATTAAAAGAATCCGGTCAGGTGCGTCATCTCGGAGTTTCGAATTTGAATTTAGAACAACTGACTAGACTCAGTAAAATCGAGAAGCCAGCTTGCTTACAAGTAGAAGTTCACCTGCTGTGCCAGCAAAAGGCTTTAGTCGAGGCTGCTAAAAAATTATCAATACCTATTGTGGCGTACTCTCCGCTCGGATCAAAAGCGCTCGCGGACGCCTTAGCTGCTAAAACTGGGCGTGATTACCCGGATCTGCTACAGCTGCCGGTAGTGAAACGTATTGCAGAGGCACACGATCGCAGCCCCGCACAGGTGCTACTCCGATATACGGTGCAACGTGGACTGGCTGCTATACCAAAAAGCACTAATCCTACTCGTATTAAACAGAACTTATCTATTTGGGATTTCGAATTAAGCGATTCCGAAATGAAAGACCTCGGTGCTTTAGACAAAGGAGAGGAGGGACGTATATGTGATTTCACATTCTTCCCAGGTATAGAGAAACATGCAGAGTTTCCATTTAAAAACTAA
Protein
MSVPEYFELNGGDRMPKIGFGTWQSSDEVLEKAVLAALDAGYRHFDTAHAYENEAALGRALRKWIGDNPAKRRELFVVTKLPPGGNRPELVREYFDTSFRNLGLDYIDMYLMHTPFAFEHVPGDLHPKNPDGTMKVDYSTDLAAVWKTLLKLKESGQVRHLGVSNLNLEQLTRLSKIEKPACLQVEVHLLCQQKALVEAAKKLSIPIVAYSPLGSKALADALAAKTGRDYPDLLQLPVVKRIAEAHDRSPAQVLLRYTVQRGLAAIPKSTNPTRIKQNLSIWDFELSDSEMKDLGALDKGEEGRICDFTFFPGIEKHAEFPFKN
Summary
Uniprot
A0A0F7QID7
A0A0Y0DEJ8
A0A076FRN3
A0A0F6Q2K7
A0A0F6Q4K1
A0A2A4IZX9
+ More
A0A2H1W0M7 A0A194RQ04 A0A194PH71 A0A310S6U0 B4KBR6 D6WTU2 Q29A01 A0A1W4VLR5 A0A2J7QCW6 B4PQY2 B4G3J8 B3NYU4 B4MBK5 A0A1Y1MCE5 B4I502 Q9VHX4 B4R0C7 B3LWW4 B4NH68 B4JYB1 A0A3B0K3W7 A0A0M4F4R0 B4MBK4 A0A2J7Q2J5 A0A1I8Q3N3 N6TN57 A0A0C9PLG1 U4UH71 T1PGH5 A0A1I8ME91 A0A087ZZ66 K7J1I4 A0A232FI94 A0A1B6LQZ9 A0A0K8V5N1 A0A182QB38 A0A034VKK9 A0A0L0BN22 A0A0K8W9C1 B4N8I1 A0A1B6IGG8 A0A195CLU3 Q7QDK4 A0A182HJR2 A0A067REM7 A0A182KWY7 A0A1B6BZ52 A0A182V201 A0A182XK76 A0A1B6G2N2 A0A0A9YK38 A0A182R804 K7J405 A0A182IP00 E2BWT3 A0A195CK08 A0A0L7QTL5 A0A026WFT0 A0A182Y4I9 Q17G72 A0A158NAF6 A0A1W4VZP2 A0A182M2M9 A0A1B0FNR1 B4NH67 A0A182W0L0 A0A023ENA0 A0A1I8P5U1 A0A1S4F3W3 A0A195EVL4 A0A182NKU4 A0A232F739 A0A1A9V8U1 K7INV3 A0A232EVU0 W8ATZ8 B4N8I0 A0A1B0A7U0 A0A2S2NSU9 H9J6A6 D3TRK6 A0A023EPE5 A0A1Q3FNX6 A0A2H8TUP4 A0A164ZMP8 A0A0N0BJQ0 A0A067QPZ1 A0A0A1WJJ7 U4UQF7 E2A7P0 A0A0M3QX16 A0A1A9XM90
A0A2H1W0M7 A0A194RQ04 A0A194PH71 A0A310S6U0 B4KBR6 D6WTU2 Q29A01 A0A1W4VLR5 A0A2J7QCW6 B4PQY2 B4G3J8 B3NYU4 B4MBK5 A0A1Y1MCE5 B4I502 Q9VHX4 B4R0C7 B3LWW4 B4NH68 B4JYB1 A0A3B0K3W7 A0A0M4F4R0 B4MBK4 A0A2J7Q2J5 A0A1I8Q3N3 N6TN57 A0A0C9PLG1 U4UH71 T1PGH5 A0A1I8ME91 A0A087ZZ66 K7J1I4 A0A232FI94 A0A1B6LQZ9 A0A0K8V5N1 A0A182QB38 A0A034VKK9 A0A0L0BN22 A0A0K8W9C1 B4N8I1 A0A1B6IGG8 A0A195CLU3 Q7QDK4 A0A182HJR2 A0A067REM7 A0A182KWY7 A0A1B6BZ52 A0A182V201 A0A182XK76 A0A1B6G2N2 A0A0A9YK38 A0A182R804 K7J405 A0A182IP00 E2BWT3 A0A195CK08 A0A0L7QTL5 A0A026WFT0 A0A182Y4I9 Q17G72 A0A158NAF6 A0A1W4VZP2 A0A182M2M9 A0A1B0FNR1 B4NH67 A0A182W0L0 A0A023ENA0 A0A1I8P5U1 A0A1S4F3W3 A0A195EVL4 A0A182NKU4 A0A232F739 A0A1A9V8U1 K7INV3 A0A232EVU0 W8ATZ8 B4N8I0 A0A1B0A7U0 A0A2S2NSU9 H9J6A6 D3TRK6 A0A023EPE5 A0A1Q3FNX6 A0A2H8TUP4 A0A164ZMP8 A0A0N0BJQ0 A0A067QPZ1 A0A0A1WJJ7 U4UQF7 E2A7P0 A0A0M3QX16 A0A1A9XM90
Pubmed
25803580
26778204
24817326
28756777
26354079
17994087
+ More
18362917 19820115 15632085 17550304 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 25315136 20075255 28648823 25348373 26108605 12364791 14747013 17210077 24845553 20966253 25401762 20798317 24508170 25244985 17510324 21347285 24945155 24495485 19121390 20353571 25830018
18362917 19820115 15632085 17550304 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 25315136 20075255 28648823 25348373 26108605 12364791 14747013 17210077 24845553 20966253 25401762 20798317 24508170 25244985 17510324 21347285 24945155 24495485 19121390 20353571 25830018
EMBL
LC017749
BAR64763.1
KU215374
AMB19668.1
KF960801
AII22003.1
+ More
KP237883 KZ149994 AKD01736.1 PZC75483.1 KP237900 AKD01753.1 NWSH01004251 PCG65301.1 ODYU01005613 SOQ46639.1 KQ460045 KPJ18106.1 KQ459604 KPI92398.1 KQ767242 OAD53474.1 CH933806 EDW14743.1 KQ971352 EFA07327.1 CM000070 EAL27550.2 NEVH01015858 PNF26431.1 CM000160 EDW96306.1 CH479179 EDW25006.1 CH954181 EDV48207.1 CH940656 EDW58476.1 GEZM01038692 JAV81556.1 CH480821 EDW55295.1 AE014297 AY118937 AAF54175.1 AAM50797.1 CM000364 EDX12052.1 CH902617 EDV42752.1 CH964272 EDW84565.1 CH916377 EDV90673.1 OUUW01000005 SPP80326.1 CP012526 ALC46787.1 EDW58475.2 NEVH01019370 PNF22802.1 APGK01058321 APGK01058322 KB741288 ENN70660.1 GBYB01001898 JAG71665.1 KB632322 ERL92352.1 KA647907 AFP62536.1 NNAY01000155 OXU30474.1 GEBQ01013874 JAT26103.1 GDHF01018123 JAI34191.1 AXCN02000306 GAKP01016609 GAKP01016607 GAKP01016604 GAKP01016602 GAKP01016599 GAKP01016596 GAKP01016594 JAC42345.1 JRES01001623 KNC21421.1 GDHF01011203 GDHF01004637 JAI41111.1 JAI47677.1 CH964232 EDW81432.1 GECU01021697 JAS86009.1 KQ977642 KYN01044.1 AAAB01008851 EAA07379.5 APCN01002207 KK852683 KDR18534.1 GEDC01030755 GEDC01024677 JAS06543.1 JAS12621.1 GECZ01013087 JAS56682.1 GBHO01011065 JAG32539.1 AAZX01001010 GL451188 EFN79797.1 KYN01048.1 KQ414754 KOC61816.1 KK107231 EZA54970.1 CH477265 EAT45576.1 ADTU01010273 AXCM01000236 CCAG010016865 EDW84564.1 GAPW01002726 JAC10872.1 KQ981954 KYN32181.1 NNAY01000838 OXU26253.1 NNAY01001957 OXU22457.1 GAMC01016868 GAMC01016861 JAB89687.1 EDW81431.1 GGMR01007652 MBY20271.1 BABH01019866 EZ424058 ADD20334.1 GAPW01002727 JAC10871.1 GFDL01005859 JAV29186.1 GFXV01006169 MBW17974.1 LRGB01000704 KZS16524.1 KQ435711 KOX79461.1 KK853074 KDR11767.1 GBXI01015260 JAC99031.1 ERL92351.1 GL437365 EFN70561.1 CP012525 ALC45079.1
KP237883 KZ149994 AKD01736.1 PZC75483.1 KP237900 AKD01753.1 NWSH01004251 PCG65301.1 ODYU01005613 SOQ46639.1 KQ460045 KPJ18106.1 KQ459604 KPI92398.1 KQ767242 OAD53474.1 CH933806 EDW14743.1 KQ971352 EFA07327.1 CM000070 EAL27550.2 NEVH01015858 PNF26431.1 CM000160 EDW96306.1 CH479179 EDW25006.1 CH954181 EDV48207.1 CH940656 EDW58476.1 GEZM01038692 JAV81556.1 CH480821 EDW55295.1 AE014297 AY118937 AAF54175.1 AAM50797.1 CM000364 EDX12052.1 CH902617 EDV42752.1 CH964272 EDW84565.1 CH916377 EDV90673.1 OUUW01000005 SPP80326.1 CP012526 ALC46787.1 EDW58475.2 NEVH01019370 PNF22802.1 APGK01058321 APGK01058322 KB741288 ENN70660.1 GBYB01001898 JAG71665.1 KB632322 ERL92352.1 KA647907 AFP62536.1 NNAY01000155 OXU30474.1 GEBQ01013874 JAT26103.1 GDHF01018123 JAI34191.1 AXCN02000306 GAKP01016609 GAKP01016607 GAKP01016604 GAKP01016602 GAKP01016599 GAKP01016596 GAKP01016594 JAC42345.1 JRES01001623 KNC21421.1 GDHF01011203 GDHF01004637 JAI41111.1 JAI47677.1 CH964232 EDW81432.1 GECU01021697 JAS86009.1 KQ977642 KYN01044.1 AAAB01008851 EAA07379.5 APCN01002207 KK852683 KDR18534.1 GEDC01030755 GEDC01024677 JAS06543.1 JAS12621.1 GECZ01013087 JAS56682.1 GBHO01011065 JAG32539.1 AAZX01001010 GL451188 EFN79797.1 KYN01048.1 KQ414754 KOC61816.1 KK107231 EZA54970.1 CH477265 EAT45576.1 ADTU01010273 AXCM01000236 CCAG010016865 EDW84564.1 GAPW01002726 JAC10872.1 KQ981954 KYN32181.1 NNAY01000838 OXU26253.1 NNAY01001957 OXU22457.1 GAMC01016868 GAMC01016861 JAB89687.1 EDW81431.1 GGMR01007652 MBY20271.1 BABH01019866 EZ424058 ADD20334.1 GAPW01002727 JAC10871.1 GFDL01005859 JAV29186.1 GFXV01006169 MBW17974.1 LRGB01000704 KZS16524.1 KQ435711 KOX79461.1 KK853074 KDR11767.1 GBXI01015260 JAC99031.1 ERL92351.1 GL437365 EFN70561.1 CP012525 ALC45079.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000009192
UP000007266
UP000001819
+ More
UP000192221 UP000235965 UP000002282 UP000008744 UP000008711 UP000008792 UP000001292 UP000000803 UP000000304 UP000007801 UP000007798 UP000001070 UP000268350 UP000092553 UP000095300 UP000019118 UP000030742 UP000095301 UP000005203 UP000002358 UP000215335 UP000075886 UP000037069 UP000078542 UP000007062 UP000075840 UP000027135 UP000075882 UP000075903 UP000076407 UP000075900 UP000075880 UP000008237 UP000053825 UP000053097 UP000076408 UP000008820 UP000005205 UP000075883 UP000092444 UP000075920 UP000078541 UP000075884 UP000078200 UP000092445 UP000005204 UP000076858 UP000053105 UP000000311 UP000092443
UP000192221 UP000235965 UP000002282 UP000008744 UP000008711 UP000008792 UP000001292 UP000000803 UP000000304 UP000007801 UP000007798 UP000001070 UP000268350 UP000092553 UP000095300 UP000019118 UP000030742 UP000095301 UP000005203 UP000002358 UP000215335 UP000075886 UP000037069 UP000078542 UP000007062 UP000075840 UP000027135 UP000075882 UP000075903 UP000076407 UP000075900 UP000075880 UP000008237 UP000053825 UP000053097 UP000076408 UP000008820 UP000005205 UP000075883 UP000092444 UP000075920 UP000078541 UP000075884 UP000078200 UP000092445 UP000005204 UP000076858 UP000053105 UP000000311 UP000092443
PRIDE
Pfam
PF00248 Aldo_ket_red
Interpro
SUPFAM
SSF51430
SSF51430
Gene 3D
ProteinModelPortal
A0A0F7QID7
A0A0Y0DEJ8
A0A076FRN3
A0A0F6Q2K7
A0A0F6Q4K1
A0A2A4IZX9
+ More
A0A2H1W0M7 A0A194RQ04 A0A194PH71 A0A310S6U0 B4KBR6 D6WTU2 Q29A01 A0A1W4VLR5 A0A2J7QCW6 B4PQY2 B4G3J8 B3NYU4 B4MBK5 A0A1Y1MCE5 B4I502 Q9VHX4 B4R0C7 B3LWW4 B4NH68 B4JYB1 A0A3B0K3W7 A0A0M4F4R0 B4MBK4 A0A2J7Q2J5 A0A1I8Q3N3 N6TN57 A0A0C9PLG1 U4UH71 T1PGH5 A0A1I8ME91 A0A087ZZ66 K7J1I4 A0A232FI94 A0A1B6LQZ9 A0A0K8V5N1 A0A182QB38 A0A034VKK9 A0A0L0BN22 A0A0K8W9C1 B4N8I1 A0A1B6IGG8 A0A195CLU3 Q7QDK4 A0A182HJR2 A0A067REM7 A0A182KWY7 A0A1B6BZ52 A0A182V201 A0A182XK76 A0A1B6G2N2 A0A0A9YK38 A0A182R804 K7J405 A0A182IP00 E2BWT3 A0A195CK08 A0A0L7QTL5 A0A026WFT0 A0A182Y4I9 Q17G72 A0A158NAF6 A0A1W4VZP2 A0A182M2M9 A0A1B0FNR1 B4NH67 A0A182W0L0 A0A023ENA0 A0A1I8P5U1 A0A1S4F3W3 A0A195EVL4 A0A182NKU4 A0A232F739 A0A1A9V8U1 K7INV3 A0A232EVU0 W8ATZ8 B4N8I0 A0A1B0A7U0 A0A2S2NSU9 H9J6A6 D3TRK6 A0A023EPE5 A0A1Q3FNX6 A0A2H8TUP4 A0A164ZMP8 A0A0N0BJQ0 A0A067QPZ1 A0A0A1WJJ7 U4UQF7 E2A7P0 A0A0M3QX16 A0A1A9XM90
A0A2H1W0M7 A0A194RQ04 A0A194PH71 A0A310S6U0 B4KBR6 D6WTU2 Q29A01 A0A1W4VLR5 A0A2J7QCW6 B4PQY2 B4G3J8 B3NYU4 B4MBK5 A0A1Y1MCE5 B4I502 Q9VHX4 B4R0C7 B3LWW4 B4NH68 B4JYB1 A0A3B0K3W7 A0A0M4F4R0 B4MBK4 A0A2J7Q2J5 A0A1I8Q3N3 N6TN57 A0A0C9PLG1 U4UH71 T1PGH5 A0A1I8ME91 A0A087ZZ66 K7J1I4 A0A232FI94 A0A1B6LQZ9 A0A0K8V5N1 A0A182QB38 A0A034VKK9 A0A0L0BN22 A0A0K8W9C1 B4N8I1 A0A1B6IGG8 A0A195CLU3 Q7QDK4 A0A182HJR2 A0A067REM7 A0A182KWY7 A0A1B6BZ52 A0A182V201 A0A182XK76 A0A1B6G2N2 A0A0A9YK38 A0A182R804 K7J405 A0A182IP00 E2BWT3 A0A195CK08 A0A0L7QTL5 A0A026WFT0 A0A182Y4I9 Q17G72 A0A158NAF6 A0A1W4VZP2 A0A182M2M9 A0A1B0FNR1 B4NH67 A0A182W0L0 A0A023ENA0 A0A1I8P5U1 A0A1S4F3W3 A0A195EVL4 A0A182NKU4 A0A232F739 A0A1A9V8U1 K7INV3 A0A232EVU0 W8ATZ8 B4N8I0 A0A1B0A7U0 A0A2S2NSU9 H9J6A6 D3TRK6 A0A023EPE5 A0A1Q3FNX6 A0A2H8TUP4 A0A164ZMP8 A0A0N0BJQ0 A0A067QPZ1 A0A0A1WJJ7 U4UQF7 E2A7P0 A0A0M3QX16 A0A1A9XM90
PDB
5KET
E-value=1.98504e-72,
Score=692
Ontologies
PATHWAY
GO
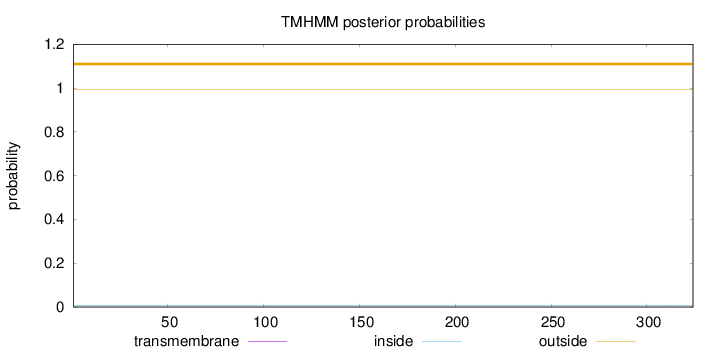
Topology
Length:
324
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00057
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00519
outside
1 - 324
Population Genetic Test Statistics
Pi
4.210875
Theta
17.161121
Tajima's D
-1.008923
CLR
192.120181
CSRT
0.135643217839108
Interpretation
Uncertain
