Pre Gene Modal
BGIBMGA005044
Annotation
PREDICTED:_HSPB1-associated_protein_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.072 Nuclear Reliability : 1.685
Sequence
CDS
ATGTCTACTATGAACGAAGACATTGTTCGCAACATAGTTGTAACAGCCAACAAACCAATAGTATTTAGAAAATTTGTCAATACTTGGCCACTAAGCAGTTGGAATGTTGAGAAATGGACCACAATATTCGGTAATAAGGAAATACCATTTCGCTGTATGAAAAAAGACTTCAGATCGGACGAACCTTGTTGGGAGCGTAGATGTGAGGTCAAAAGTATGACTTTCAAGCAGTTTATAGATAATTTGGACACCAGCGAAGAGTGGATGTACTTCGACTACAAATACGTCCATAAATGGTTTGTAAGCGAGGATGAACTTTATAAGGACATATCATGGAAGGATTTTGGTTTTCCAAATAAAGGAGCGTCAGATACTACTTTGTGGATTGGCAGCAAGGGCTCACACACACCAGCACACCAAGACACATATGGAGTTAATATTGTGGCACAATTATATGGAAAGAAGCGTTGGATACTGTTCCCCCCAGAAACCGGTGGATTAAAACCGACTAGGGTGCCTTATGAAGAGTCAAGTATTTACAGTGAATTAAATTTCCATTGTCCAAGCGATTTGGAGCAATTTAAGCACTTGACTGGAGCTAGAGTAGTGGATCTGACTGCAGGTGATGCGCTTGTGGTACCCAGGGCATGGTGGCATTACGTACAAAATATGGATCCTCTCAACATATCTCTGAATACCTGGCTACCTCATGAAAAAGATACATCAACTCAGGTGTCGGAAGCTCTCATCAAAATATTTGTAGCACAGATATGTAAGGATCTTCCTGAAGATACAGCAAAATTGTTGATAAATCCGAATGAGGATGATATATCGGTGACGCCTTTGGTCGTGCTGTTCCTGCAGCTAGAGATGGTCGCCAAAAGTTATTTGGATAACCGACGTAAAACGCGACGCGCCAAGCGACAAAAGACCTGCGAAGAGAATCAATCGGCAACCGAAAGCGGAGAACTCAATATTGAGGCTCTCATACAAAACGAGTGTAATAAGCTGGAGGTACCGCCGCCGATAACTGGCGAACAACTTGTGGACATGATCAGACAAAATCTGCGCGAATATGTTAATTTCGAACGGCCTTTACACGACGACGAGATTGATGGATCGACTTCAAGCCTTTGTTTAACTAAAGCCCTTATAGACGCGTTCACGCAGAGCAATGTCATAGATATTGTAAAGGAGAATTTATTTGCCAGACTAAGCTAA
Protein
MSTMNEDIVRNIVVTANKPIVFRKFVNTWPLSSWNVEKWTTIFGNKEIPFRCMKKDFRSDEPCWERRCEVKSMTFKQFIDNLDTSEEWMYFDYKYVHKWFVSEDELYKDISWKDFGFPNKGASDTTLWIGSKGSHTPAHQDTYGVNIVAQLYGKKRWILFPPETGGLKPTRVPYEESSIYSELNFHCPSDLEQFKHLTGARVVDLTAGDALVVPRAWWHYVQNMDPLNISLNTWLPHEKDTSTQVSEALIKIFVAQICKDLPEDTAKLLINPNEDDISVTPLVVLFLQLEMVAKSYLDNRRKTRRAKRQKTCEENQSATESGELNIEALIQNECNKLEVPPPITGEQLVDMIRQNLREYVNFERPLHDDEIDGSTSSLCLTKALIDAFTQSNVIDIVKENLFARLS
Summary
Uniprot
H9J6A3
A0A2A4J272
A0A2W1BPI8
A0A3S2MAS8
A0A194PG82
A0A212FJ90
+ More
A0A194RL14 A0A0L7LP51 A0A2J7QXQ5 A0A067RDC7 N6ST03 J3JWA5 A0A1W4XDW3 A0A069DSR2 A0A0P5HR84 A0A0P6HEX5 A0A0P6AGA1 A0A091WHR3 A0A162SE06 A0A091RM19 A0A2D4PEN5 A0A093PUY0 A0A1V4K7F3 A0A091N210 A0A091I496 A0A2I0MG89 A0A093GP01 W5M337 V8NWP1 A0A3Q2EI53 H3DE34 A0A091LXN1 A0A091SNF7 A0A091KGW6 R0LM80 H0Z1T7 A0A087V9U3 A0A3P9IE03 G1KDF0 U3I8C1 A0A3P9MFF7 A0A2U9CBS8 A0A0P7U4A6 A0A1W5A624 A0A1S3JUW7 A0A1A8LLN9 A0A1A8ICR2 A0A1A7ZAK1 A0A1A8CMP2 A0A1A8QHJ4 A0A091MXN9 A0A3B3DND0 A0A0P4YSJ6
A0A194RL14 A0A0L7LP51 A0A2J7QXQ5 A0A067RDC7 N6ST03 J3JWA5 A0A1W4XDW3 A0A069DSR2 A0A0P5HR84 A0A0P6HEX5 A0A0P6AGA1 A0A091WHR3 A0A162SE06 A0A091RM19 A0A2D4PEN5 A0A093PUY0 A0A1V4K7F3 A0A091N210 A0A091I496 A0A2I0MG89 A0A093GP01 W5M337 V8NWP1 A0A3Q2EI53 H3DE34 A0A091LXN1 A0A091SNF7 A0A091KGW6 R0LM80 H0Z1T7 A0A087V9U3 A0A3P9IE03 G1KDF0 U3I8C1 A0A3P9MFF7 A0A2U9CBS8 A0A0P7U4A6 A0A1W5A624 A0A1S3JUW7 A0A1A8LLN9 A0A1A8ICR2 A0A1A7ZAK1 A0A1A8CMP2 A0A1A8QHJ4 A0A091MXN9 A0A3B3DND0 A0A0P4YSJ6
Pubmed
EMBL
BABH01019869
NWSH01003770
PCG65836.1
KZ149994
PZC75485.1
RSAL01000001
+ More
RVE55321.1 KQ459604 KPI92396.1 AGBW02008295 OWR53808.1 KQ460045 KPJ18109.1 JTDY01000432 KOB77200.1 NEVH01009372 PNF33367.1 KK852540 KDR21767.1 APGK01057851 APGK01057852 APGK01057853 APGK01057854 KB741282 ENN70824.1 BT127523 AEE62485.1 GBGD01002177 JAC86712.1 GDIQ01226053 JAK25672.1 GDIQ01020425 JAN74312.1 GDIP01031145 JAM72570.1 KK733897 KFR01116.1 LRGB01000044 KZS21211.1 KK821589 KFQ40302.1 IACN01063345 LAB56495.1 KL670757 KFW80196.1 LSYS01004331 OPJ79827.1 KK840301 KFP82929.1 KL218243 KFP03364.1 AKCR02000014 PKK28709.1 KK368809 KFV42435.1 AHAT01011609 AZIM01001553 ETE66470.1 KL299837 KFP51066.1 KK929706 KFQ44562.1 KK744737 KFP39834.1 KB742929 EOB02800.1 ABQF01010986 KL485229 KFO09385.1 ADON01058481 CP026256 AWP13490.1 JARO02009170 KPP61916.1 HAEF01007432 SBR44814.1 HAED01008854 SBQ95066.1 HADY01000655 HAEJ01007413 SBP39140.1 HADZ01017083 SBP81024.1 HAEG01012645 SBR92887.1 KL372444 KFP81442.1 GDIP01233870 JAI89531.1
RVE55321.1 KQ459604 KPI92396.1 AGBW02008295 OWR53808.1 KQ460045 KPJ18109.1 JTDY01000432 KOB77200.1 NEVH01009372 PNF33367.1 KK852540 KDR21767.1 APGK01057851 APGK01057852 APGK01057853 APGK01057854 KB741282 ENN70824.1 BT127523 AEE62485.1 GBGD01002177 JAC86712.1 GDIQ01226053 JAK25672.1 GDIQ01020425 JAN74312.1 GDIP01031145 JAM72570.1 KK733897 KFR01116.1 LRGB01000044 KZS21211.1 KK821589 KFQ40302.1 IACN01063345 LAB56495.1 KL670757 KFW80196.1 LSYS01004331 OPJ79827.1 KK840301 KFP82929.1 KL218243 KFP03364.1 AKCR02000014 PKK28709.1 KK368809 KFV42435.1 AHAT01011609 AZIM01001553 ETE66470.1 KL299837 KFP51066.1 KK929706 KFQ44562.1 KK744737 KFP39834.1 KB742929 EOB02800.1 ABQF01010986 KL485229 KFO09385.1 ADON01058481 CP026256 AWP13490.1 JARO02009170 KPP61916.1 HAEF01007432 SBR44814.1 HAED01008854 SBQ95066.1 HADY01000655 HAEJ01007413 SBP39140.1 HADZ01017083 SBP81024.1 HAEG01012645 SBR92887.1 KL372444 KFP81442.1 GDIP01233870 JAI89531.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
+ More
UP000037510 UP000235965 UP000027135 UP000019118 UP000192223 UP000053605 UP000076858 UP000053258 UP000190648 UP000054308 UP000053872 UP000018468 UP000265020 UP000007303 UP000007754 UP000265200 UP000001646 UP000016666 UP000265180 UP000246464 UP000034805 UP000192224 UP000085678 UP000261560
UP000037510 UP000235965 UP000027135 UP000019118 UP000192223 UP000053605 UP000076858 UP000053258 UP000190648 UP000054308 UP000053872 UP000018468 UP000265020 UP000007303 UP000007754 UP000265200 UP000001646 UP000016666 UP000265180 UP000246464 UP000034805 UP000192224 UP000085678 UP000261560
PRIDE
Interpro
SUPFAM
SSF158573
SSF158573
ProteinModelPortal
H9J6A3
A0A2A4J272
A0A2W1BPI8
A0A3S2MAS8
A0A194PG82
A0A212FJ90
+ More
A0A194RL14 A0A0L7LP51 A0A2J7QXQ5 A0A067RDC7 N6ST03 J3JWA5 A0A1W4XDW3 A0A069DSR2 A0A0P5HR84 A0A0P6HEX5 A0A0P6AGA1 A0A091WHR3 A0A162SE06 A0A091RM19 A0A2D4PEN5 A0A093PUY0 A0A1V4K7F3 A0A091N210 A0A091I496 A0A2I0MG89 A0A093GP01 W5M337 V8NWP1 A0A3Q2EI53 H3DE34 A0A091LXN1 A0A091SNF7 A0A091KGW6 R0LM80 H0Z1T7 A0A087V9U3 A0A3P9IE03 G1KDF0 U3I8C1 A0A3P9MFF7 A0A2U9CBS8 A0A0P7U4A6 A0A1W5A624 A0A1S3JUW7 A0A1A8LLN9 A0A1A8ICR2 A0A1A7ZAK1 A0A1A8CMP2 A0A1A8QHJ4 A0A091MXN9 A0A3B3DND0 A0A0P4YSJ6
A0A194RL14 A0A0L7LP51 A0A2J7QXQ5 A0A067RDC7 N6ST03 J3JWA5 A0A1W4XDW3 A0A069DSR2 A0A0P5HR84 A0A0P6HEX5 A0A0P6AGA1 A0A091WHR3 A0A162SE06 A0A091RM19 A0A2D4PEN5 A0A093PUY0 A0A1V4K7F3 A0A091N210 A0A091I496 A0A2I0MG89 A0A093GP01 W5M337 V8NWP1 A0A3Q2EI53 H3DE34 A0A091LXN1 A0A091SNF7 A0A091KGW6 R0LM80 H0Z1T7 A0A087V9U3 A0A3P9IE03 G1KDF0 U3I8C1 A0A3P9MFF7 A0A2U9CBS8 A0A0P7U4A6 A0A1W5A624 A0A1S3JUW7 A0A1A8LLN9 A0A1A8ICR2 A0A1A7ZAK1 A0A1A8CMP2 A0A1A8QHJ4 A0A091MXN9 A0A3B3DND0 A0A0P4YSJ6
PDB
6F4Q
E-value=1.98345e-12,
Score=175
Ontologies
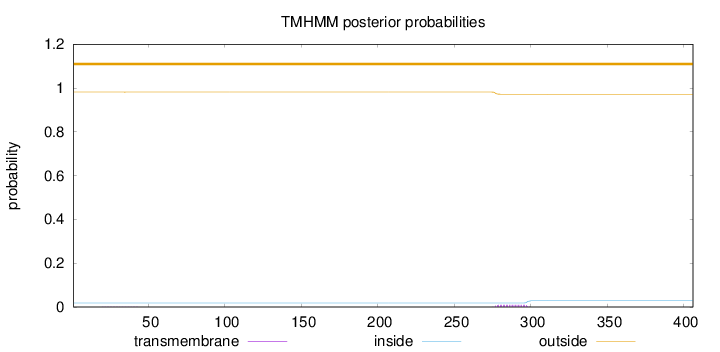
Topology
Subcellular location
Nucleus
Length:
406
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21765
Exp number, first 60 AAs:
0.00632
Total prob of N-in:
0.01848
outside
1 - 406
Population Genetic Test Statistics
Pi
161.721513
Theta
169.278188
Tajima's D
-0.866931
CLR
176.320614
CSRT
0.16279186040698
Interpretation
Uncertain
