Pre Gene Modal
BGIBMGA005158
Annotation
trafficking_protein_particle_complex_6b_[Bombyx_mori]
Full name
DNA polymerase alpha subunit B
Location in the cell
PlasmaMembrane Reliability : 1.942
Sequence
CDS
ATGGCCGACGACATCATATTTGAATTATTACATTCTGAAATTATAAATTATTCTATTGAAAAATTCAAGAATAACGATAATGGCGAAAAGGAAACTGATTTGTCTGTTGTCGAATACATCGGGTTTGCGGCAGGCTATAAAATCATGGAAAGATTAACGAGGGAATGGCCGCGTTTTAAAGATGAATTAGATACTATGAAATTCATATGTACAGATTTTTGGACTTGTATTTACAAAAAACAGATAGATAATCTTCGTACAAACCACCAAGGAGTTTATGTACTTCAAGATAATGCATTCCGTTTTCTAACAAATTTTTCTAATGGTCATCAGTACCTGGAATATGCTCCTAGATATGTTGCTTACACTTGTGGGCTCATTCGAGGTGGATTGGCTAATCTCTGTATTAACAGTATTGTCACAGCAGAAGTTCAGTCTATGCCTTCATGTAAATTTCATATACACGTTCAAAGAACATAA
Protein
MADDIIFELLHSEIINYSIEKFKNNDNGEKETDLSVVEYIGFAAGYKIMERLTREWPRFKDELDTMKFICTDFWTCIYKKQIDNLRTNHQGVYVLQDNAFRFLTNFSNGHQYLEYAPRYVAYTCGLIRGGLANLCINSIVTAEVQSMPSCKFHIHVQRT
Summary
Description
May play an essential role at the early stage of chromosomal DNA replication by coupling the polymerase alpha/primase complex to the cellular replication machinery.
Similarity
Belongs to the DNA polymerase alpha subunit B family.
Uniprot
Q1HPK2
A0A2W1BKC0
S4PSA6
A0A1E1WI46
A0A3S2P2B6
A0A194PH68
+ More
A0A194RKP7 A0A212FJA2 A0A2H1WZ21 A0A0L7LH06 A0A2J7QRY4 A0A1I8JWA6 A0A2C9GUL6 A0A1Y9H9M4 A0A1Y9IS63 A0A1C7CZV4 A0A240PM17 A0A1I8JV53 A0A1I8JTK3 A0A182K0N5 Q7QC03 A0A182RYF3 A0A240PMM8 A0A1Y9IVB0 A0A1Y9H301 A0A182Y771 A0A182SHZ8 V5FW02 A0A1W4XKP5 A0A1Y9G9Q3 T1DP14 A0A2M3ZBZ7 A0A2M4AR04 A0A1Q3FKA0 B0WF96 A0A2M4CMK3 A0A067REY7 Q17E67 A0A087Z0L9 A0A023EFH7 A0A2M4AQZ7 W5JQ82 U5EW98 N6U752 A0A1Y1NCN3 H9J6L6 A0A0K8TP59 A0A1B0CSJ7 A0A1L8DVZ2 A0A1B0DI74 U5EUE0 A0A1B6DIG2 A0A1B6M7R5 A0A1B6CGQ7 A0A1B6JC29 A0A1B6GJH5 A0A0L0BND8 A0A0A9WDS9 B3MSU3 B4NAT8 D3TRF6 A0A240SX99 A0A1B6DC05 A0A0K8VDB9 A0A1W4VTS6 A0A1A9W7C9 A0A1B6DML6 A0A1J1HIX1 A0A3B0K414 Q29BE4 B4GNU2 A0A2R7W936 A0A1A9VG02 A0A1I8MF74 B4HM25 R4FNN0 B4QYI6 B4PS42 B3P0V6 Q9VF82 B4JU44 A0A195CY59 E2ATA3 A0A1I8PBM2 B4M5Z7 A0A0J7KZ18 A0A069DW92 A0A1B0B2Q7 E2C0B9 A0A026WIH2 A0A336KFM6 E0V955 A0A224XVD7 A0A240SWM7 B4KAQ3 A0A0M3QYE7 A0A2C9JJ75 A0A195E6Q8 A0A195B1T0 A0A195F1M0
A0A194RKP7 A0A212FJA2 A0A2H1WZ21 A0A0L7LH06 A0A2J7QRY4 A0A1I8JWA6 A0A2C9GUL6 A0A1Y9H9M4 A0A1Y9IS63 A0A1C7CZV4 A0A240PM17 A0A1I8JV53 A0A1I8JTK3 A0A182K0N5 Q7QC03 A0A182RYF3 A0A240PMM8 A0A1Y9IVB0 A0A1Y9H301 A0A182Y771 A0A182SHZ8 V5FW02 A0A1W4XKP5 A0A1Y9G9Q3 T1DP14 A0A2M3ZBZ7 A0A2M4AR04 A0A1Q3FKA0 B0WF96 A0A2M4CMK3 A0A067REY7 Q17E67 A0A087Z0L9 A0A023EFH7 A0A2M4AQZ7 W5JQ82 U5EW98 N6U752 A0A1Y1NCN3 H9J6L6 A0A0K8TP59 A0A1B0CSJ7 A0A1L8DVZ2 A0A1B0DI74 U5EUE0 A0A1B6DIG2 A0A1B6M7R5 A0A1B6CGQ7 A0A1B6JC29 A0A1B6GJH5 A0A0L0BND8 A0A0A9WDS9 B3MSU3 B4NAT8 D3TRF6 A0A240SX99 A0A1B6DC05 A0A0K8VDB9 A0A1W4VTS6 A0A1A9W7C9 A0A1B6DML6 A0A1J1HIX1 A0A3B0K414 Q29BE4 B4GNU2 A0A2R7W936 A0A1A9VG02 A0A1I8MF74 B4HM25 R4FNN0 B4QYI6 B4PS42 B3P0V6 Q9VF82 B4JU44 A0A195CY59 E2ATA3 A0A1I8PBM2 B4M5Z7 A0A0J7KZ18 A0A069DW92 A0A1B0B2Q7 E2C0B9 A0A026WIH2 A0A336KFM6 E0V955 A0A224XVD7 A0A240SWM7 B4KAQ3 A0A0M3QYE7 A0A2C9JJ75 A0A195E6Q8 A0A195B1T0 A0A195F1M0
Pubmed
28756777
23622113
26354079
22118469
26227816
20966253
+ More
12364791 14747013 17210077 25244985 24845553 17510324 20920257 24945155 26483478 23537049 28004739 19121390 26369729 26108605 25401762 26823975 17994087 20353571 15632085 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 26334808 24508170 30249741 20566863 15562597
12364791 14747013 17210077 25244985 24845553 17510324 20920257 24945155 26483478 23537049 28004739 19121390 26369729 26108605 25401762 26823975 17994087 20353571 15632085 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 26334808 24508170 30249741 20566863 15562597
EMBL
DQ443400
ABF51489.1
KZ149994
PZC75489.1
GAIX01000105
JAA92455.1
+ More
GDQN01004426 JAT86628.1 RSAL01000001 RVE55323.1 KQ459604 KPI92393.1 KQ460045 KPJ18112.1 AGBW02008295 OWR53812.1 ODYU01012138 SOQ58278.1 JTDY01001220 KOB74491.1 NEVH01011882 PNF31348.1 AXCM01000066 AXCN02000218 APCN01000423 AAAB01008859 EAA08080.2 GALX01006626 JAB61840.1 GAMD01002365 JAA99225.1 GGFM01005323 MBW26074.1 GGFK01009879 MBW43200.1 GFDL01007139 JAV27906.1 DS231916 EDS26113.1 GGFL01002366 MBW66544.1 KK852730 KDR17521.1 CH477285 EAT44752.1 JXUM01135890 GAPW01005426 GAPW01005425 KQ568348 JAC08173.1 KXJ69037.1 GGFK01009880 MBW43201.1 ADMH02000650 ETN65448.1 GANO01001518 JAB58353.1 APGK01036917 KB740945 KB632197 ENN77480.1 ERL89997.1 GEZM01008998 JAV94660.1 BABH01019869 GDAI01001655 JAI15948.1 AJWK01026149 GFDF01003488 JAV10596.1 AJVK01062114 GANO01001519 JAB58352.1 GEDC01011870 JAS25428.1 GEBQ01008019 JAT31958.1 GEDC01024773 JAS12525.1 GECU01011048 JAS96658.1 GECZ01011429 GECZ01007336 JAS58340.1 JAS62433.1 JRES01001623 KNC21438.1 GBHO01038064 GBHO01038063 GBRD01005878 GDHC01021324 GDHC01015968 JAG05540.1 JAG05541.1 JAG59943.1 JAP97304.1 JAQ02661.1 CH902623 EDV30333.1 CH964232 EDW80902.2 EZ424008 ADD20284.1 GEDC01014084 JAS23214.1 GDHF01015375 JAI36939.1 GEDC01010391 JAS26907.1 CVRI01000006 CRK88000.1 OUUW01000005 SPP80739.1 CM000070 EAL27054.1 CH479186 EDW38825.1 KK854406 PTY15560.1 CH480815 EDW42062.1 GAHY01001224 JAA76286.1 CM000364 EDX12831.1 CM000160 EDW97462.1 CH954181 EDV49004.1 AE014297 AY071721 KX532055 AAF55178.1 AAL49343.1 ANY27865.1 CH916374 EDV91014.1 KQ977115 KYN05605.1 GL442545 EFN63348.1 CH940652 EDW59073.1 LBMM01001916 KMQ95548.1 GBGD01003295 JAC85594.1 JXJN01007687 GL451770 EFN78578.1 KK107185 QOIP01000012 EZA55815.1 RLU15906.1 UFQS01000309 UFQT01000309 SSX02712.1 SSX23086.1 DS234988 EEB09911.1 GFTR01002628 JAW13798.1 CH933806 EDW16790.2 CP012526 ALC47489.1 KQ979568 KYN20776.1 KQ976662 KYM78431.1 KQ981864 KYN34363.1
GDQN01004426 JAT86628.1 RSAL01000001 RVE55323.1 KQ459604 KPI92393.1 KQ460045 KPJ18112.1 AGBW02008295 OWR53812.1 ODYU01012138 SOQ58278.1 JTDY01001220 KOB74491.1 NEVH01011882 PNF31348.1 AXCM01000066 AXCN02000218 APCN01000423 AAAB01008859 EAA08080.2 GALX01006626 JAB61840.1 GAMD01002365 JAA99225.1 GGFM01005323 MBW26074.1 GGFK01009879 MBW43200.1 GFDL01007139 JAV27906.1 DS231916 EDS26113.1 GGFL01002366 MBW66544.1 KK852730 KDR17521.1 CH477285 EAT44752.1 JXUM01135890 GAPW01005426 GAPW01005425 KQ568348 JAC08173.1 KXJ69037.1 GGFK01009880 MBW43201.1 ADMH02000650 ETN65448.1 GANO01001518 JAB58353.1 APGK01036917 KB740945 KB632197 ENN77480.1 ERL89997.1 GEZM01008998 JAV94660.1 BABH01019869 GDAI01001655 JAI15948.1 AJWK01026149 GFDF01003488 JAV10596.1 AJVK01062114 GANO01001519 JAB58352.1 GEDC01011870 JAS25428.1 GEBQ01008019 JAT31958.1 GEDC01024773 JAS12525.1 GECU01011048 JAS96658.1 GECZ01011429 GECZ01007336 JAS58340.1 JAS62433.1 JRES01001623 KNC21438.1 GBHO01038064 GBHO01038063 GBRD01005878 GDHC01021324 GDHC01015968 JAG05540.1 JAG05541.1 JAG59943.1 JAP97304.1 JAQ02661.1 CH902623 EDV30333.1 CH964232 EDW80902.2 EZ424008 ADD20284.1 GEDC01014084 JAS23214.1 GDHF01015375 JAI36939.1 GEDC01010391 JAS26907.1 CVRI01000006 CRK88000.1 OUUW01000005 SPP80739.1 CM000070 EAL27054.1 CH479186 EDW38825.1 KK854406 PTY15560.1 CH480815 EDW42062.1 GAHY01001224 JAA76286.1 CM000364 EDX12831.1 CM000160 EDW97462.1 CH954181 EDV49004.1 AE014297 AY071721 KX532055 AAF55178.1 AAL49343.1 ANY27865.1 CH916374 EDV91014.1 KQ977115 KYN05605.1 GL442545 EFN63348.1 CH940652 EDW59073.1 LBMM01001916 KMQ95548.1 GBGD01003295 JAC85594.1 JXJN01007687 GL451770 EFN78578.1 KK107185 QOIP01000012 EZA55815.1 RLU15906.1 UFQS01000309 UFQT01000309 SSX02712.1 SSX23086.1 DS234988 EEB09911.1 GFTR01002628 JAW13798.1 CH933806 EDW16790.2 CP012526 ALC47489.1 KQ979568 KYN20776.1 KQ976662 KYM78431.1 KQ981864 KYN34363.1
Proteomes
UP000283053
UP000053268
UP000053240
UP000007151
UP000037510
UP000235965
+ More
UP000076407 UP000075883 UP000075886 UP000075903 UP000075882 UP000075885 UP000075902 UP000075840 UP000075881 UP000007062 UP000075900 UP000075880 UP000075920 UP000075884 UP000076408 UP000075901 UP000192223 UP000069272 UP000002320 UP000027135 UP000008820 UP000069940 UP000249989 UP000000673 UP000019118 UP000030742 UP000005204 UP000092461 UP000092462 UP000037069 UP000007801 UP000007798 UP000092445 UP000192221 UP000091820 UP000183832 UP000268350 UP000001819 UP000008744 UP000078200 UP000095301 UP000001292 UP000000304 UP000002282 UP000008711 UP000000803 UP000001070 UP000078542 UP000000311 UP000095300 UP000008792 UP000036403 UP000092460 UP000008237 UP000053097 UP000279307 UP000009046 UP000092443 UP000009192 UP000092553 UP000076420 UP000078492 UP000078540 UP000078541
UP000076407 UP000075883 UP000075886 UP000075903 UP000075882 UP000075885 UP000075902 UP000075840 UP000075881 UP000007062 UP000075900 UP000075880 UP000075920 UP000075884 UP000076408 UP000075901 UP000192223 UP000069272 UP000002320 UP000027135 UP000008820 UP000069940 UP000249989 UP000000673 UP000019118 UP000030742 UP000005204 UP000092461 UP000092462 UP000037069 UP000007801 UP000007798 UP000092445 UP000192221 UP000091820 UP000183832 UP000268350 UP000001819 UP000008744 UP000078200 UP000095301 UP000001292 UP000000304 UP000002282 UP000008711 UP000000803 UP000001070 UP000078542 UP000000311 UP000095300 UP000008792 UP000036403 UP000092460 UP000008237 UP000053097 UP000279307 UP000009046 UP000092443 UP000009192 UP000092553 UP000076420 UP000078492 UP000078540 UP000078541
Interpro
SUPFAM
SSF111126
SSF111126
ProteinModelPortal
Q1HPK2
A0A2W1BKC0
S4PSA6
A0A1E1WI46
A0A3S2P2B6
A0A194PH68
+ More
A0A194RKP7 A0A212FJA2 A0A2H1WZ21 A0A0L7LH06 A0A2J7QRY4 A0A1I8JWA6 A0A2C9GUL6 A0A1Y9H9M4 A0A1Y9IS63 A0A1C7CZV4 A0A240PM17 A0A1I8JV53 A0A1I8JTK3 A0A182K0N5 Q7QC03 A0A182RYF3 A0A240PMM8 A0A1Y9IVB0 A0A1Y9H301 A0A182Y771 A0A182SHZ8 V5FW02 A0A1W4XKP5 A0A1Y9G9Q3 T1DP14 A0A2M3ZBZ7 A0A2M4AR04 A0A1Q3FKA0 B0WF96 A0A2M4CMK3 A0A067REY7 Q17E67 A0A087Z0L9 A0A023EFH7 A0A2M4AQZ7 W5JQ82 U5EW98 N6U752 A0A1Y1NCN3 H9J6L6 A0A0K8TP59 A0A1B0CSJ7 A0A1L8DVZ2 A0A1B0DI74 U5EUE0 A0A1B6DIG2 A0A1B6M7R5 A0A1B6CGQ7 A0A1B6JC29 A0A1B6GJH5 A0A0L0BND8 A0A0A9WDS9 B3MSU3 B4NAT8 D3TRF6 A0A240SX99 A0A1B6DC05 A0A0K8VDB9 A0A1W4VTS6 A0A1A9W7C9 A0A1B6DML6 A0A1J1HIX1 A0A3B0K414 Q29BE4 B4GNU2 A0A2R7W936 A0A1A9VG02 A0A1I8MF74 B4HM25 R4FNN0 B4QYI6 B4PS42 B3P0V6 Q9VF82 B4JU44 A0A195CY59 E2ATA3 A0A1I8PBM2 B4M5Z7 A0A0J7KZ18 A0A069DW92 A0A1B0B2Q7 E2C0B9 A0A026WIH2 A0A336KFM6 E0V955 A0A224XVD7 A0A240SWM7 B4KAQ3 A0A0M3QYE7 A0A2C9JJ75 A0A195E6Q8 A0A195B1T0 A0A195F1M0
A0A194RKP7 A0A212FJA2 A0A2H1WZ21 A0A0L7LH06 A0A2J7QRY4 A0A1I8JWA6 A0A2C9GUL6 A0A1Y9H9M4 A0A1Y9IS63 A0A1C7CZV4 A0A240PM17 A0A1I8JV53 A0A1I8JTK3 A0A182K0N5 Q7QC03 A0A182RYF3 A0A240PMM8 A0A1Y9IVB0 A0A1Y9H301 A0A182Y771 A0A182SHZ8 V5FW02 A0A1W4XKP5 A0A1Y9G9Q3 T1DP14 A0A2M3ZBZ7 A0A2M4AR04 A0A1Q3FKA0 B0WF96 A0A2M4CMK3 A0A067REY7 Q17E67 A0A087Z0L9 A0A023EFH7 A0A2M4AQZ7 W5JQ82 U5EW98 N6U752 A0A1Y1NCN3 H9J6L6 A0A0K8TP59 A0A1B0CSJ7 A0A1L8DVZ2 A0A1B0DI74 U5EUE0 A0A1B6DIG2 A0A1B6M7R5 A0A1B6CGQ7 A0A1B6JC29 A0A1B6GJH5 A0A0L0BND8 A0A0A9WDS9 B3MSU3 B4NAT8 D3TRF6 A0A240SX99 A0A1B6DC05 A0A0K8VDB9 A0A1W4VTS6 A0A1A9W7C9 A0A1B6DML6 A0A1J1HIX1 A0A3B0K414 Q29BE4 B4GNU2 A0A2R7W936 A0A1A9VG02 A0A1I8MF74 B4HM25 R4FNN0 B4QYI6 B4PS42 B3P0V6 Q9VF82 B4JU44 A0A195CY59 E2ATA3 A0A1I8PBM2 B4M5Z7 A0A0J7KZ18 A0A069DW92 A0A1B0B2Q7 E2C0B9 A0A026WIH2 A0A336KFM6 E0V955 A0A224XVD7 A0A240SWM7 B4KAQ3 A0A0M3QYE7 A0A2C9JJ75 A0A195E6Q8 A0A195B1T0 A0A195F1M0
PDB
2BJN
E-value=2.20712e-51,
Score=505
Ontologies
GO
Topology
Subcellular location
Nucleus
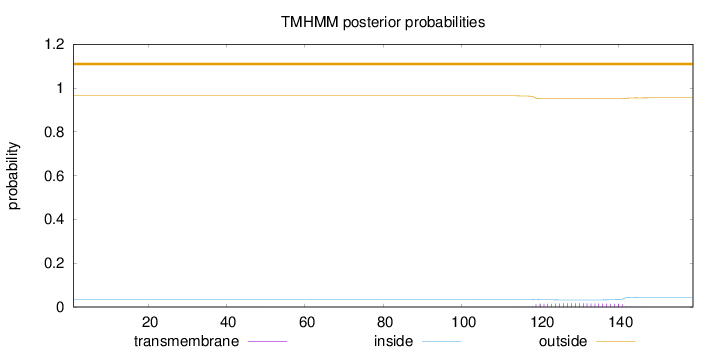
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.36754
Exp number, first 60 AAs:
0.00319
Total prob of N-in:
0.03523
outside
1 - 159
Population Genetic Test Statistics
Pi
156.601539
Theta
164.542204
Tajima's D
-0.172935
CLR
23.773523
CSRT
0.321933903304835
Interpretation
Uncertain
