Pre Gene Modal
BGIBMGA005158
Annotation
PREDICTED:_DNA_polymerase_alpha_subunit_B_[Papilio_polytes]
Full name
DNA polymerase alpha subunit B
Alternative Name
DNA polymerase alpha 70 kDa subunit
Location in the cell
Extracellular Reliability : 1.112 Nuclear Reliability : 1.367
Sequence
CDS
ATGGCCTCCGAAGAGTTGGTGACTGAGCAATTTAAATTTTTTGGCATAAACGTTGAACAAGATGTTTTATTAAAATGTGTAAATTTATGTGAAGAGAACAATATTGATGCTGAAACTTTTATCGAACAATGGATGGCATTCAGTTTGAATTATGTAAACGGTGCTTCACCAAACTTGGAAAACTTAGATTTATTCGTAAGAAAGGAATTTTCCAAGCGCCGAACTAACGCGTCCGCTAAAGAAACCACACGACCAGCTGCCGGTAAAAGTCTTACCGTTTATGGAGCAAAAGCCACTTCGCCTCCAGACAGTGAAGTACTGTCAAGTTATGTTGCCGTAACGCCCAAGAGAGTCAAAATCGAAAAAGACGCGACGACAAATCCGAATAACATAGTTCCTGCAACTTATTCACCGACCGTCGGTTCAGCGAAGTACGCGTCGAGAACAAACGCAGGCACGGTGGTCCATTCTTATGGAGATGAGAAATTACTACAAGAATTAGGTAAAGCGAACAGCTCGAATGATGTGCTCAGTTTGAAAATCACGCAAGTGCCTAATGACGATGACGAAGTTTACACAAAGGCGATGTTGGGTTTTGAACTTCTCCACGAAAAAGCCAGTACGTTTGATAATCACATTCGATACATATCGCAGTACATAATGAAGAAGGCGGGTGTAAAGGAAAGTACCTCAGTTAGACACAAAACACAGACAAACGTATACGTTGCTGGACGTATAGAATGTGATGCAGATGCGAGGTTAAATGCAAAATGTGTGGTGCTTCAAGGTACGTGGGAGGAGTCACTCAGTCAGTCGGTACCGGTGGACTTTGACAATGTATATCAGTATTCCTTATTCCCTGGTCAAGTGGTGGTGCTGCGCGGAATAAATCCCCGCGGTGATAGGTTTATAGCACAAGAAGTTTATTGTGACGCAATCCAACCATTACCTGACCATAAGTTCGATATAGCCGGTAAATTGTCGATGGTCGCTGTGGCGGGCCCGTACACGACGTCGGACAACATGGCCTACGAACCTCTCAAGGACATGGTGACGTACATAAACACGCACAAACCGCACGTAGTCATAATGACGGGGCCGTTTATTGACAGTGAACACTCCAAAGTCAAAGACAATACGATGGCCGAGACATTTAGCTCGTTGTTTGGGAAAATTCTCGATAGTCTGGGAGAATTGAGCAACACGAGTCCGGTCACGAAAATCTATATCGTATCGAATTTGAAGGACGCTTTCCATGTTAATGTCTACCCCACGCCGCCGTACGTGAGCCGAAGGAAACACCCCAACATACATTTTGTACCGGATCCCTGCACTCTTACCATTAATGGTATCCACGTGGGAGTTACTAGTACAGATATCCTCATGCACATTAGCCAGGAAGAAATATCACTAGGCACCGGCGGCGACAAGCTGTCCCGCTTAGCGAGCCACGTGCTGTCGCAGCAGACGTACTACCCGCTGTGTCCGGCGCCCGTCCCGCTGGACGCCGCGCTGTGGGCCGCGCACGCACAGCTTCAGGCATCTCCGCATCTGCTCGTACTACCCTCCAATTTCAGATATTTTGTTAAGGATGTCAACGGATGTGTAGTTGTCAATCCAGAACATTTAACAAAAGGCACCGGCGGAGGGACCTTCACTCGTGTATTAATCGATAGCGACAACGCCACGAAACGAATTGCAGCACAAATTGTCCGCATTTAA
Protein
MASEELVTEQFKFFGINVEQDVLLKCVNLCEENNIDAETFIEQWMAFSLNYVNGASPNLENLDLFVRKEFSKRRTNASAKETTRPAAGKSLTVYGAKATSPPDSEVLSSYVAVTPKRVKIEKDATTNPNNIVPATYSPTVGSAKYASRTNAGTVVHSYGDEKLLQELGKANSSNDVLSLKITQVPNDDDEVYTKAMLGFELLHEKASTFDNHIRYISQYIMKKAGVKESTSVRHKTQTNVYVAGRIECDADARLNAKCVVLQGTWEESLSQSVPVDFDNVYQYSLFPGQVVVLRGINPRGDRFIAQEVYCDAIQPLPDHKFDIAGKLSMVAVAGPYTTSDNMAYEPLKDMVTYINTHKPHVVIMTGPFIDSEHSKVKDNTMAETFSSLFGKILDSLGELSNTSPVTKIYIVSNLKDAFHVNVYPTPPYVSRRKHPNIHFVPDPCTLTINGIHVGVTSTDILMHISQEEISLGTGGDKLSRLASHVLSQQTYYPLCPAPVPLDAALWAAHAQLQASPHLLVLPSNFRYFVKDVNGCVVVNPEHLTKGTGGGTFTRVLIDSDNATKRIAAQIVRI
Summary
Description
May play an essential role at the early stage of chromosomal DNA replication by coupling the polymerase alpha/primase complex to the cellular replication machinery.
Subunit
DNA polymerase alpha:primase is a four subunit enzyme complex, which is assembled throughout the cell cycle, and consists of the two DNA polymerase subunits A and B, and the DNA primase large and small subunits. Subunit B binds to subunit A.
Similarity
Belongs to the DNA polymerase alpha subunit B family.
Keywords
3D-structure
Alternative splicing
Complete proteome
DNA replication
Nucleus
Phosphoprotein
Polymorphism
Reference proteome
Feature
chain DNA polymerase alpha subunit B
splice variant In isoform 2.
sequence variant In dbSNP:rs487989.
splice variant In isoform 2.
sequence variant In dbSNP:rs487989.
Uniprot
H9J6L6
A0A2W1BRT2
A0A2H1WZ21
A0A212FJ97
A0A194RL19
A0A2A4J3A7
+ More
A0A0L7LH06 A0A067RAB1 A0A2J7QE41 A0A1B6FG21 A0A1B6IGW6 A0A2J7QE45 A0A1Y1KDB5 V4BAC9 A0A1S3KAP8 A0A2C9KJI2 K1QC65 A0A3Q1B401 A0A0P4YM17 A0A0P5SBQ0 A0A0P5T8G3 A0A0P5JWK6 A0A1B6I9E5 A0A0P5ZC15 A0A0P5U1H3 A0A0P5UWM6 A0A3Q1GQ62 A0A0P6DXJ9 A0A0P5ZUI8 A0A3Q1H2G5 A0A0P6G296 A0A0P5GDL7 Q6DCZ1 A0A0P5QYW1 A0A0N8D2B4 A0A0P6DWS8 A0A3B4XTK1 A0A0P5KCV0 A0A3P8SRS3 A0A0P5U0R4 A0A0P5YIT1 A0A0K8UDF0 A0A0P5LFC7 Q6DEY9 A0A3B4UV79 A0A1L8GJG1 A0A0P6E102 A0A0K8VVQ6 A0A0P5KPI2 A0A2K6NHA0 Q28C83 A0A0P5QYR9 A0A0P6FQT0 A0A0N8AC26 A0A131YQB6 A0A0P5HRH2 A0A1A8GFA3 A0A1S3FKI4 A0A3P8V2I6 A0A3Q1IDT2 A0A2J8TZ97 A0A0P5TKX2 A0A1A8I5X5 A0A2Y9PPQ4 A0A0P5BBU8 Q7ZVB6 A0A293LR25 A0A3Q3JP60 A0A340XPG9 H2Q426 A0A1A8SGG2 G3QKN0 A0A1A8QBU2 A0A2K6EC81 Q14181 A0A3P8Q138 A0A3B3V0V0 B0S687 A0A3P9B644 K7B9Q6 A0A341BQ96 A0A1Y1KGH6 A0A250Y3E0 Q6P0S9 A0A3Q3KHA9 A0A383Z747 A0A1A7WRM5 A0A087XZJ9 A0A2K5DY80 H0XBR5 A0A0N8A356 A0A0P5TH20 A0A2Y9PV55 A0A3B3WHP3 A0A2G8KDD6 A0A2K5KER4
A0A0L7LH06 A0A067RAB1 A0A2J7QE41 A0A1B6FG21 A0A1B6IGW6 A0A2J7QE45 A0A1Y1KDB5 V4BAC9 A0A1S3KAP8 A0A2C9KJI2 K1QC65 A0A3Q1B401 A0A0P4YM17 A0A0P5SBQ0 A0A0P5T8G3 A0A0P5JWK6 A0A1B6I9E5 A0A0P5ZC15 A0A0P5U1H3 A0A0P5UWM6 A0A3Q1GQ62 A0A0P6DXJ9 A0A0P5ZUI8 A0A3Q1H2G5 A0A0P6G296 A0A0P5GDL7 Q6DCZ1 A0A0P5QYW1 A0A0N8D2B4 A0A0P6DWS8 A0A3B4XTK1 A0A0P5KCV0 A0A3P8SRS3 A0A0P5U0R4 A0A0P5YIT1 A0A0K8UDF0 A0A0P5LFC7 Q6DEY9 A0A3B4UV79 A0A1L8GJG1 A0A0P6E102 A0A0K8VVQ6 A0A0P5KPI2 A0A2K6NHA0 Q28C83 A0A0P5QYR9 A0A0P6FQT0 A0A0N8AC26 A0A131YQB6 A0A0P5HRH2 A0A1A8GFA3 A0A1S3FKI4 A0A3P8V2I6 A0A3Q1IDT2 A0A2J8TZ97 A0A0P5TKX2 A0A1A8I5X5 A0A2Y9PPQ4 A0A0P5BBU8 Q7ZVB6 A0A293LR25 A0A3Q3JP60 A0A340XPG9 H2Q426 A0A1A8SGG2 G3QKN0 A0A1A8QBU2 A0A2K6EC81 Q14181 A0A3P8Q138 A0A3B3V0V0 B0S687 A0A3P9B644 K7B9Q6 A0A341BQ96 A0A1Y1KGH6 A0A250Y3E0 Q6P0S9 A0A3Q3KHA9 A0A383Z747 A0A1A7WRM5 A0A087XZJ9 A0A2K5DY80 H0XBR5 A0A0N8A356 A0A0P5TH20 A0A2Y9PV55 A0A3B3WHP3 A0A2G8KDD6 A0A2K5KER4
Pubmed
19121390
28756777
22118469
26354079
26227816
24845553
+ More
28004739 23254933 15562597 22992520 20431018 27762356 25362486 26830274 24487278 16136131 22398555 8223465 14702039 16554811 15489334 1902230 18669648 19413330 19690332 20068231 21269460 21406692 23186163 23594743 25186727 28087693 29023486
28004739 23254933 15562597 22992520 20431018 27762356 25362486 26830274 24487278 16136131 22398555 8223465 14702039 16554811 15489334 1902230 18669648 19413330 19690332 20068231 21269460 21406692 23186163 23594743 25186727 28087693 29023486
EMBL
BABH01019869
KZ149994
PZC75490.1
ODYU01012138
SOQ58278.1
AGBW02008295
+ More
OWR53813.1 KQ460045 KPJ18114.1 NWSH01003726 PCG65883.1 JTDY01001220 KOB74491.1 KK852590 KDR20724.1 NEVH01015328 PNF26859.1 GECZ01020613 JAS49156.1 GECU01021548 GECU01017099 JAS86158.1 JAS90607.1 PNF26860.1 GEZM01086413 JAV59469.1 KB200129 ESP02897.1 JH817954 EKC28759.1 GDIP01225380 JAI98021.1 GDIQ01094238 JAL57488.1 GDIQ01203914 GDIP01131789 GDIQ01029930 JAK47811.1 JAL71925.1 GDIQ01192457 JAK59268.1 GECU01024142 GECU01005906 JAS83564.1 JAT01801.1 GDIP01058706 JAM45009.1 GDIP01121332 JAL82382.1 GDIP01107787 JAL95927.1 GDIQ01070799 JAN23938.1 GDIP01039403 JAM64312.1 GDIQ01039558 JAN55179.1 GDIQ01249749 GDIQ01186556 GDIQ01121321 GDIQ01067658 JAK01976.1 BC077841 AAH77841.1 GDIQ01108292 JAL43434.1 GDIQ01203913 GDIQ01203912 GDIQ01144080 GDIP01075735 JAK47812.1 JAM27980.1 GDIQ01071653 JAN23084.1 GDIQ01192456 JAK59269.1 GDIP01121333 JAL82381.1 GDIP01058707 JAM45008.1 GDHF01027622 JAI24692.1 GDIQ01170423 JAK81302.1 AAMC01070934 BC076954 AAH76954.1 CM004472 OCT83985.1 GDIQ01070798 JAN23939.1 GDHF01009352 JAI42962.1 GDIQ01186557 GDIQ01186555 GDIQ01158118 GDIQ01121320 GDIQ01067659 JAK65168.1 CR942408 CAJ82570.1 GDIQ01108337 JAL43389.1 GDIQ01044559 JAN50178.1 GDIP01162129 JAJ61273.1 GEDV01007769 JAP80788.1 GDIQ01229748 JAK21977.1 HAEB01013914 HAEC01000987 SBQ69064.1 NDHI03003478 PNJ38347.1 GDIP01126340 JAL77374.1 HAED01006241 SBQ92272.1 GDIP01187090 JAJ36312.1 BC045928 AAH45928.1 GFWV01005581 MAA30311.1 AACZ04016215 AACZ04016216 GABF01008635 JAA13510.1 HAEH01015582 HAEI01014534 SBS17003.1 CABD030079717 CABD030079718 CABD030079719 HAEF01013133 HAEG01011844 SBR90802.1 L24559 AK297842 CR456737 AP000944 BC002990 BC001347 BX465215 GABC01007951 GABD01007067 GABE01007284 NBAG03000050 JAA03387.1 JAA26033.1 JAA37455.1 PNI93838.1 GEZM01086412 JAV59471.1 GFFV01001839 JAV38106.1 BC065460 AAH65460.1 HADW01006993 SBP08393.1 AYCK01003857 AAQR03147612 AAQR03147613 AAQR03147614 AAQR03147615 AAQR03147616 AAQR03147617 AAQR03147618 AAQR03147619 AAQR03147620 GDIP01187089 JAJ36313.1 GDIP01126339 JAL77375.1 MRZV01000670 PIK46017.1
OWR53813.1 KQ460045 KPJ18114.1 NWSH01003726 PCG65883.1 JTDY01001220 KOB74491.1 KK852590 KDR20724.1 NEVH01015328 PNF26859.1 GECZ01020613 JAS49156.1 GECU01021548 GECU01017099 JAS86158.1 JAS90607.1 PNF26860.1 GEZM01086413 JAV59469.1 KB200129 ESP02897.1 JH817954 EKC28759.1 GDIP01225380 JAI98021.1 GDIQ01094238 JAL57488.1 GDIQ01203914 GDIP01131789 GDIQ01029930 JAK47811.1 JAL71925.1 GDIQ01192457 JAK59268.1 GECU01024142 GECU01005906 JAS83564.1 JAT01801.1 GDIP01058706 JAM45009.1 GDIP01121332 JAL82382.1 GDIP01107787 JAL95927.1 GDIQ01070799 JAN23938.1 GDIP01039403 JAM64312.1 GDIQ01039558 JAN55179.1 GDIQ01249749 GDIQ01186556 GDIQ01121321 GDIQ01067658 JAK01976.1 BC077841 AAH77841.1 GDIQ01108292 JAL43434.1 GDIQ01203913 GDIQ01203912 GDIQ01144080 GDIP01075735 JAK47812.1 JAM27980.1 GDIQ01071653 JAN23084.1 GDIQ01192456 JAK59269.1 GDIP01121333 JAL82381.1 GDIP01058707 JAM45008.1 GDHF01027622 JAI24692.1 GDIQ01170423 JAK81302.1 AAMC01070934 BC076954 AAH76954.1 CM004472 OCT83985.1 GDIQ01070798 JAN23939.1 GDHF01009352 JAI42962.1 GDIQ01186557 GDIQ01186555 GDIQ01158118 GDIQ01121320 GDIQ01067659 JAK65168.1 CR942408 CAJ82570.1 GDIQ01108337 JAL43389.1 GDIQ01044559 JAN50178.1 GDIP01162129 JAJ61273.1 GEDV01007769 JAP80788.1 GDIQ01229748 JAK21977.1 HAEB01013914 HAEC01000987 SBQ69064.1 NDHI03003478 PNJ38347.1 GDIP01126340 JAL77374.1 HAED01006241 SBQ92272.1 GDIP01187090 JAJ36312.1 BC045928 AAH45928.1 GFWV01005581 MAA30311.1 AACZ04016215 AACZ04016216 GABF01008635 JAA13510.1 HAEH01015582 HAEI01014534 SBS17003.1 CABD030079717 CABD030079718 CABD030079719 HAEF01013133 HAEG01011844 SBR90802.1 L24559 AK297842 CR456737 AP000944 BC002990 BC001347 BX465215 GABC01007951 GABD01007067 GABE01007284 NBAG03000050 JAA03387.1 JAA26033.1 JAA37455.1 PNI93838.1 GEZM01086412 JAV59471.1 GFFV01001839 JAV38106.1 BC065460 AAH65460.1 HADW01006993 SBP08393.1 AYCK01003857 AAQR03147612 AAQR03147613 AAQR03147614 AAQR03147615 AAQR03147616 AAQR03147617 AAQR03147618 AAQR03147619 AAQR03147620 GDIP01187089 JAJ36313.1 GDIP01126339 JAL77375.1 MRZV01000670 PIK46017.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000218220
UP000037510
UP000027135
+ More
UP000235965 UP000030746 UP000085678 UP000076420 UP000005408 UP000257160 UP000257200 UP000261360 UP000265080 UP000008143 UP000261420 UP000186698 UP000233200 UP000081671 UP000265120 UP000265040 UP000248483 UP000261600 UP000265300 UP000002277 UP000001519 UP000233120 UP000005640 UP000265100 UP000261500 UP000000437 UP000265160 UP000252040 UP000261640 UP000261681 UP000028760 UP000233020 UP000005225 UP000261480 UP000230750 UP000233080
UP000235965 UP000030746 UP000085678 UP000076420 UP000005408 UP000257160 UP000257200 UP000261360 UP000265080 UP000008143 UP000261420 UP000186698 UP000233200 UP000081671 UP000265120 UP000265040 UP000248483 UP000261600 UP000265300 UP000002277 UP000001519 UP000233120 UP000005640 UP000265100 UP000261500 UP000000437 UP000265160 UP000252040 UP000261640 UP000261681 UP000028760 UP000233020 UP000005225 UP000261480 UP000230750 UP000233080
Interpro
SUPFAM
SSF111126
SSF111126
ProteinModelPortal
H9J6L6
A0A2W1BRT2
A0A2H1WZ21
A0A212FJ97
A0A194RL19
A0A2A4J3A7
+ More
A0A0L7LH06 A0A067RAB1 A0A2J7QE41 A0A1B6FG21 A0A1B6IGW6 A0A2J7QE45 A0A1Y1KDB5 V4BAC9 A0A1S3KAP8 A0A2C9KJI2 K1QC65 A0A3Q1B401 A0A0P4YM17 A0A0P5SBQ0 A0A0P5T8G3 A0A0P5JWK6 A0A1B6I9E5 A0A0P5ZC15 A0A0P5U1H3 A0A0P5UWM6 A0A3Q1GQ62 A0A0P6DXJ9 A0A0P5ZUI8 A0A3Q1H2G5 A0A0P6G296 A0A0P5GDL7 Q6DCZ1 A0A0P5QYW1 A0A0N8D2B4 A0A0P6DWS8 A0A3B4XTK1 A0A0P5KCV0 A0A3P8SRS3 A0A0P5U0R4 A0A0P5YIT1 A0A0K8UDF0 A0A0P5LFC7 Q6DEY9 A0A3B4UV79 A0A1L8GJG1 A0A0P6E102 A0A0K8VVQ6 A0A0P5KPI2 A0A2K6NHA0 Q28C83 A0A0P5QYR9 A0A0P6FQT0 A0A0N8AC26 A0A131YQB6 A0A0P5HRH2 A0A1A8GFA3 A0A1S3FKI4 A0A3P8V2I6 A0A3Q1IDT2 A0A2J8TZ97 A0A0P5TKX2 A0A1A8I5X5 A0A2Y9PPQ4 A0A0P5BBU8 Q7ZVB6 A0A293LR25 A0A3Q3JP60 A0A340XPG9 H2Q426 A0A1A8SGG2 G3QKN0 A0A1A8QBU2 A0A2K6EC81 Q14181 A0A3P8Q138 A0A3B3V0V0 B0S687 A0A3P9B644 K7B9Q6 A0A341BQ96 A0A1Y1KGH6 A0A250Y3E0 Q6P0S9 A0A3Q3KHA9 A0A383Z747 A0A1A7WRM5 A0A087XZJ9 A0A2K5DY80 H0XBR5 A0A0N8A356 A0A0P5TH20 A0A2Y9PV55 A0A3B3WHP3 A0A2G8KDD6 A0A2K5KER4
A0A0L7LH06 A0A067RAB1 A0A2J7QE41 A0A1B6FG21 A0A1B6IGW6 A0A2J7QE45 A0A1Y1KDB5 V4BAC9 A0A1S3KAP8 A0A2C9KJI2 K1QC65 A0A3Q1B401 A0A0P4YM17 A0A0P5SBQ0 A0A0P5T8G3 A0A0P5JWK6 A0A1B6I9E5 A0A0P5ZC15 A0A0P5U1H3 A0A0P5UWM6 A0A3Q1GQ62 A0A0P6DXJ9 A0A0P5ZUI8 A0A3Q1H2G5 A0A0P6G296 A0A0P5GDL7 Q6DCZ1 A0A0P5QYW1 A0A0N8D2B4 A0A0P6DWS8 A0A3B4XTK1 A0A0P5KCV0 A0A3P8SRS3 A0A0P5U0R4 A0A0P5YIT1 A0A0K8UDF0 A0A0P5LFC7 Q6DEY9 A0A3B4UV79 A0A1L8GJG1 A0A0P6E102 A0A0K8VVQ6 A0A0P5KPI2 A0A2K6NHA0 Q28C83 A0A0P5QYR9 A0A0P6FQT0 A0A0N8AC26 A0A131YQB6 A0A0P5HRH2 A0A1A8GFA3 A0A1S3FKI4 A0A3P8V2I6 A0A3Q1IDT2 A0A2J8TZ97 A0A0P5TKX2 A0A1A8I5X5 A0A2Y9PPQ4 A0A0P5BBU8 Q7ZVB6 A0A293LR25 A0A3Q3JP60 A0A340XPG9 H2Q426 A0A1A8SGG2 G3QKN0 A0A1A8QBU2 A0A2K6EC81 Q14181 A0A3P8Q138 A0A3B3V0V0 B0S687 A0A3P9B644 K7B9Q6 A0A341BQ96 A0A1Y1KGH6 A0A250Y3E0 Q6P0S9 A0A3Q3KHA9 A0A383Z747 A0A1A7WRM5 A0A087XZJ9 A0A2K5DY80 H0XBR5 A0A0N8A356 A0A0P5TH20 A0A2Y9PV55 A0A3B3WHP3 A0A2G8KDD6 A0A2K5KER4
PDB
5EXR
E-value=2.05184e-79,
Score=754
Ontologies
GO
PANTHER
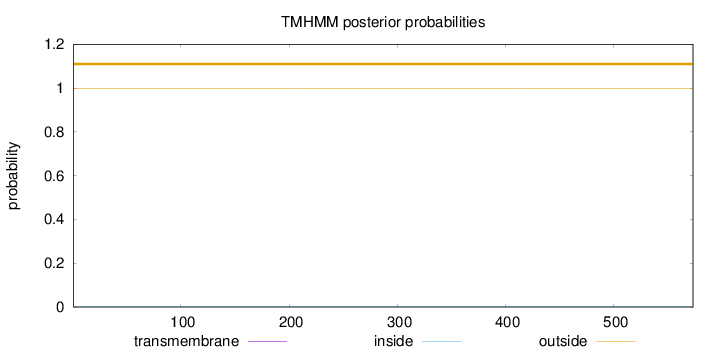
Topology
Subcellular location
Nucleus
Length:
573
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00298
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00078
outside
1 - 573
Population Genetic Test Statistics
Pi
194.678767
Theta
156.189658
Tajima's D
0.94853
CLR
1.001405
CSRT
0.650917454127294
Interpretation
Uncertain
