Pre Gene Modal
BGIBMGA005159
Annotation
signal_recognition_particle_14_kDa_protein-like_protein_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.61 Nuclear Reliability : 1.801
Sequence
CDS
ATGGTTTTATTGAGCAACGATGAGTTTCTAGCAGAGTTAACAAAACTTTTCCAAAAAGCACGATTAGCAGGATCAATCACAATGACTATGAAGAGATATGACGGCCGTAGCAAACCAATGCCTCGAGACGGAACTCCTGCAGTTAAAAATCCCGAATACAAATGCCTATTACGGGCACAATCTAAAAGTAAAAAGATATCAACAGTTGTGGAACAGCGTGATGTTGAAAAGTTCTCAACTGCATATTCTAATCTGCTGAAAACGAGTGTTAATGGCTTGAAACGCTTGAAAAAGCCCAAGAAAAAAGCAATGGCGACTCAGTAG
Protein
MVLLSNDEFLAELTKLFQKARLAGSITMTMKRYDGRSKPMPRDGTPAVKNPEYKCLLRAQSKSKKISTVVEQRDVEKFSTAYSNLLKTSVNGLKRLKKPKKKAMATQ
Summary
Uniprot
Q0N2R9
A0A1E1W449
H9J6L7
A0A2A4J2C4
A0A2H1WYW6
S4NJ47
+ More
A0A194RLC1 I4DPK1 A0A194PI35 A0A0L7LGF0 A0A212FJA6 A0A2W1BUR2 A0A1Y1LJP3 D6WT83 A0A1W4XPK6 E2A848 A0A2A3EQW8 A0A087ZZQ6 E0W0A2 A0A1Z5KXQ6 A0A158NT37 N6TN85 A0A3L8DDH0 K7J957 A0A2P8YHZ9 A0A067RK01 A0A1A7YDB1 A0A023FW76 A0A1A8HG77 A0A1A8EEY8 A0A1A8PNY3 A0A1A8M3D9 A0A1A8JTH0 A0A1A8V490 A0A1E1XRV9 A0A0N5DNE0 A0A1S3P0A8 A0A224Z8S6 A0A131Z4V4 L7M5Z6 A0A131X8X4 B7QAY0 V5I2G7 A0A060WNH9 W5UMA9 A0A1S3SNS3 A0A087UQB7 A0A2C9KHX1 A0A3B1J581 A0A3Q2E4M4 A0A060WF73 B6RB61 A0A0P7ZA42 A0A0B1PKG4 A0A3P8XEZ2 A0A077Z113 K4G058 A0A232EQP0 A0A210QWI2 A0A3Q2QGL0 A0A0K8RE66 A0A3B3WP10 A0A0S7HSN9 M3ZWP3 A0A087XVF3 A0A3B3VED6 A0A3P9QC74 S4RRY0 A0A3B3RIV1 A0A3B3CFI2 K4GIG1 A0A3B3CH02 A0A1Q3EU74 Q4V9I5 A0A250Y987 A0A085MX62 A0A1B6GLV5 C9W1F3 W5N535 G3SMS7 A0A2Y9DUE0 A0A0B7AW55 A0A3Q3LQD6 A0A3Q3DKY4 E9IBD6 A0A0N0BDG3 A0A0B7AWH3 A0A224XQC8 A0A1S4EY08 T1E3B6 A0A3Q1EW71 A0A3Q1HN29 A0A3Q1BAV6 A0A3P8RK49 A0A3B4EUU4 A0A3Q2VPQ6 A0A3Q4M3M6 I3K2H9 A0A3P8UHT0 I3MQ27
A0A194RLC1 I4DPK1 A0A194PI35 A0A0L7LGF0 A0A212FJA6 A0A2W1BUR2 A0A1Y1LJP3 D6WT83 A0A1W4XPK6 E2A848 A0A2A3EQW8 A0A087ZZQ6 E0W0A2 A0A1Z5KXQ6 A0A158NT37 N6TN85 A0A3L8DDH0 K7J957 A0A2P8YHZ9 A0A067RK01 A0A1A7YDB1 A0A023FW76 A0A1A8HG77 A0A1A8EEY8 A0A1A8PNY3 A0A1A8M3D9 A0A1A8JTH0 A0A1A8V490 A0A1E1XRV9 A0A0N5DNE0 A0A1S3P0A8 A0A224Z8S6 A0A131Z4V4 L7M5Z6 A0A131X8X4 B7QAY0 V5I2G7 A0A060WNH9 W5UMA9 A0A1S3SNS3 A0A087UQB7 A0A2C9KHX1 A0A3B1J581 A0A3Q2E4M4 A0A060WF73 B6RB61 A0A0P7ZA42 A0A0B1PKG4 A0A3P8XEZ2 A0A077Z113 K4G058 A0A232EQP0 A0A210QWI2 A0A3Q2QGL0 A0A0K8RE66 A0A3B3WP10 A0A0S7HSN9 M3ZWP3 A0A087XVF3 A0A3B3VED6 A0A3P9QC74 S4RRY0 A0A3B3RIV1 A0A3B3CFI2 K4GIG1 A0A3B3CH02 A0A1Q3EU74 Q4V9I5 A0A250Y987 A0A085MX62 A0A1B6GLV5 C9W1F3 W5N535 G3SMS7 A0A2Y9DUE0 A0A0B7AW55 A0A3Q3LQD6 A0A3Q3DKY4 E9IBD6 A0A0N0BDG3 A0A0B7AWH3 A0A224XQC8 A0A1S4EY08 T1E3B6 A0A3Q1EW71 A0A3Q1HN29 A0A3Q1BAV6 A0A3P8RK49 A0A3B4EUU4 A0A3Q2VPQ6 A0A3Q4M3M6 I3K2H9 A0A3P8UHT0 I3MQ27
Pubmed
19121390
23622113
26354079
22651552
26227816
22118469
+ More
28756777 28004739 18362917 19820115 20798317 20566863 28528879 21347285 23537049 30249741 20075255 29403074 24845553 29209593 28797301 26830274 25576852 28049606 25765539 24755649 23127152 15562597 25329095 25069045 23056606 28648823 28812685 23542700 29240929 29451363 23594743 28087693 24929829 20650005 24029695 21282665 24330624 25186727 24487278
28756777 28004739 18362917 19820115 20798317 20566863 28528879 21347285 23537049 30249741 20075255 29403074 24845553 29209593 28797301 26830274 25576852 28049606 25765539 24755649 23127152 15562597 25329095 25069045 23056606 28648823 28812685 23542700 29240929 29451363 23594743 28087693 24929829 20650005 24029695 21282665 24330624 25186727 24487278
EMBL
DQ646412
ABH10802.1
GDQN01009296
JAT81758.1
BABH01019872
NWSH01003726
+ More
PCG65886.1 ODYU01012138 SOQ58280.1 GAIX01013824 JAA78736.1 KQ460045 KPJ18115.1 AK403645 BAM19841.1 KQ459604 KPI92389.1 JTDY01001220 KOB74490.1 AGBW02008295 OWR53815.1 KZ149994 PZC75493.1 GEZM01054004 JAV73884.1 KQ971354 EFA07651.2 GL437496 EFN70393.1 KZ288194 PBC33894.1 DS235857 EEB19058.1 GFJQ02007047 JAV99922.1 ADTU01025488 APGK01058388 APGK01058389 KB741288 KB632370 ENN70695.1 ERL93728.1 QOIP01000009 RLU18515.1 AAZX01002219 PYGN01000580 PSN43885.1 KK852425 KDR24161.1 HADW01015555 HADX01006275 SBP28507.1 GBBL01002357 JAC24963.1 HAEB01006455 HAEC01015004 SBQ83221.1 HAEA01015159 SBQ43639.1 HAEH01007759 HAEI01004232 SBR82717.1 HAEF01011052 HAEG01006115 SBR51575.1 HAED01015623 HAEE01003405 SBR23425.1 HADY01005423 HAEJ01015057 SBS55514.1 GFAA01001382 JAU02053.1 GFPF01011756 MAA22902.1 GEDV01002549 JAP86008.1 GACK01006510 JAA58524.1 GEFH01004587 JAP63994.1 ABJB010519555 ABJB010921648 DS897208 EEC16002.1 GANP01000756 GEFM01001760 JAB83712.1 JAP74036.1 FR904635 CDQ68602.1 JT418787 AHH42845.1 KK121016 KFM79556.1 FR904523 CDQ65943.1 EF103407 ABO26665.1 JARO02000699 KPP77690.1 KN538445 KHJ40270.1 HG805861 CDW53629.1 JX052463 AFK10691.1 NNAY01002732 OXU20675.1 NEDP02001506 OWF53129.1 GADI01005039 JAA68769.1 GBYX01437213 JAO44134.1 AYCK01003395 JX208383 AFM86697.1 GFDL01016181 JAV18864.1 LO018432 BC096882 AAH96882.1 GFFW01004664 JAV40124.1 KL363303 KL367610 KFD48118.1 KFD61808.1 GECZ01006344 JAS63425.1 EZ406101 ACX53900.1 AHAT01019028 HACG01037396 CEK84261.1 GL762111 EFZ22053.1 KQ435863 KOX70411.1 HACG01037395 CEK84260.1 GFTR01001719 JAW14707.1 GALA01000515 JAA94337.1 AERX01018670 AGTP01002150
PCG65886.1 ODYU01012138 SOQ58280.1 GAIX01013824 JAA78736.1 KQ460045 KPJ18115.1 AK403645 BAM19841.1 KQ459604 KPI92389.1 JTDY01001220 KOB74490.1 AGBW02008295 OWR53815.1 KZ149994 PZC75493.1 GEZM01054004 JAV73884.1 KQ971354 EFA07651.2 GL437496 EFN70393.1 KZ288194 PBC33894.1 DS235857 EEB19058.1 GFJQ02007047 JAV99922.1 ADTU01025488 APGK01058388 APGK01058389 KB741288 KB632370 ENN70695.1 ERL93728.1 QOIP01000009 RLU18515.1 AAZX01002219 PYGN01000580 PSN43885.1 KK852425 KDR24161.1 HADW01015555 HADX01006275 SBP28507.1 GBBL01002357 JAC24963.1 HAEB01006455 HAEC01015004 SBQ83221.1 HAEA01015159 SBQ43639.1 HAEH01007759 HAEI01004232 SBR82717.1 HAEF01011052 HAEG01006115 SBR51575.1 HAED01015623 HAEE01003405 SBR23425.1 HADY01005423 HAEJ01015057 SBS55514.1 GFAA01001382 JAU02053.1 GFPF01011756 MAA22902.1 GEDV01002549 JAP86008.1 GACK01006510 JAA58524.1 GEFH01004587 JAP63994.1 ABJB010519555 ABJB010921648 DS897208 EEC16002.1 GANP01000756 GEFM01001760 JAB83712.1 JAP74036.1 FR904635 CDQ68602.1 JT418787 AHH42845.1 KK121016 KFM79556.1 FR904523 CDQ65943.1 EF103407 ABO26665.1 JARO02000699 KPP77690.1 KN538445 KHJ40270.1 HG805861 CDW53629.1 JX052463 AFK10691.1 NNAY01002732 OXU20675.1 NEDP02001506 OWF53129.1 GADI01005039 JAA68769.1 GBYX01437213 JAO44134.1 AYCK01003395 JX208383 AFM86697.1 GFDL01016181 JAV18864.1 LO018432 BC096882 AAH96882.1 GFFW01004664 JAV40124.1 KL363303 KL367610 KFD48118.1 KFD61808.1 GECZ01006344 JAS63425.1 EZ406101 ACX53900.1 AHAT01019028 HACG01037396 CEK84261.1 GL762111 EFZ22053.1 KQ435863 KOX70411.1 HACG01037395 CEK84260.1 GFTR01001719 JAW14707.1 GALA01000515 JAA94337.1 AERX01018670 AGTP01002150
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000007266 UP000192223 UP000000311 UP000242457 UP000005203 UP000009046 UP000005205 UP000019118 UP000030742 UP000279307 UP000002358 UP000245037 UP000027135 UP000046395 UP000087266 UP000001555 UP000193380 UP000221080 UP000054359 UP000076420 UP000018467 UP000265020 UP000034805 UP000192224 UP000265140 UP000030665 UP000215335 UP000242188 UP000265000 UP000261480 UP000002852 UP000028760 UP000261500 UP000242638 UP000245300 UP000261540 UP000261560 UP000000437 UP000018468 UP000007646 UP000248480 UP000261640 UP000264820 UP000053105 UP000257200 UP000265040 UP000257160 UP000265080 UP000261460 UP000264840 UP000261580 UP000005207 UP000265120 UP000005215
UP000007266 UP000192223 UP000000311 UP000242457 UP000005203 UP000009046 UP000005205 UP000019118 UP000030742 UP000279307 UP000002358 UP000245037 UP000027135 UP000046395 UP000087266 UP000001555 UP000193380 UP000221080 UP000054359 UP000076420 UP000018467 UP000265020 UP000034805 UP000192224 UP000265140 UP000030665 UP000215335 UP000242188 UP000265000 UP000261480 UP000002852 UP000028760 UP000261500 UP000242638 UP000245300 UP000261540 UP000261560 UP000000437 UP000018468 UP000007646 UP000248480 UP000261640 UP000264820 UP000053105 UP000257200 UP000265040 UP000257160 UP000265080 UP000261460 UP000264840 UP000261580 UP000005207 UP000265120 UP000005215
Interpro
Gene 3D
CDD
ProteinModelPortal
Q0N2R9
A0A1E1W449
H9J6L7
A0A2A4J2C4
A0A2H1WYW6
S4NJ47
+ More
A0A194RLC1 I4DPK1 A0A194PI35 A0A0L7LGF0 A0A212FJA6 A0A2W1BUR2 A0A1Y1LJP3 D6WT83 A0A1W4XPK6 E2A848 A0A2A3EQW8 A0A087ZZQ6 E0W0A2 A0A1Z5KXQ6 A0A158NT37 N6TN85 A0A3L8DDH0 K7J957 A0A2P8YHZ9 A0A067RK01 A0A1A7YDB1 A0A023FW76 A0A1A8HG77 A0A1A8EEY8 A0A1A8PNY3 A0A1A8M3D9 A0A1A8JTH0 A0A1A8V490 A0A1E1XRV9 A0A0N5DNE0 A0A1S3P0A8 A0A224Z8S6 A0A131Z4V4 L7M5Z6 A0A131X8X4 B7QAY0 V5I2G7 A0A060WNH9 W5UMA9 A0A1S3SNS3 A0A087UQB7 A0A2C9KHX1 A0A3B1J581 A0A3Q2E4M4 A0A060WF73 B6RB61 A0A0P7ZA42 A0A0B1PKG4 A0A3P8XEZ2 A0A077Z113 K4G058 A0A232EQP0 A0A210QWI2 A0A3Q2QGL0 A0A0K8RE66 A0A3B3WP10 A0A0S7HSN9 M3ZWP3 A0A087XVF3 A0A3B3VED6 A0A3P9QC74 S4RRY0 A0A3B3RIV1 A0A3B3CFI2 K4GIG1 A0A3B3CH02 A0A1Q3EU74 Q4V9I5 A0A250Y987 A0A085MX62 A0A1B6GLV5 C9W1F3 W5N535 G3SMS7 A0A2Y9DUE0 A0A0B7AW55 A0A3Q3LQD6 A0A3Q3DKY4 E9IBD6 A0A0N0BDG3 A0A0B7AWH3 A0A224XQC8 A0A1S4EY08 T1E3B6 A0A3Q1EW71 A0A3Q1HN29 A0A3Q1BAV6 A0A3P8RK49 A0A3B4EUU4 A0A3Q2VPQ6 A0A3Q4M3M6 I3K2H9 A0A3P8UHT0 I3MQ27
A0A194RLC1 I4DPK1 A0A194PI35 A0A0L7LGF0 A0A212FJA6 A0A2W1BUR2 A0A1Y1LJP3 D6WT83 A0A1W4XPK6 E2A848 A0A2A3EQW8 A0A087ZZQ6 E0W0A2 A0A1Z5KXQ6 A0A158NT37 N6TN85 A0A3L8DDH0 K7J957 A0A2P8YHZ9 A0A067RK01 A0A1A7YDB1 A0A023FW76 A0A1A8HG77 A0A1A8EEY8 A0A1A8PNY3 A0A1A8M3D9 A0A1A8JTH0 A0A1A8V490 A0A1E1XRV9 A0A0N5DNE0 A0A1S3P0A8 A0A224Z8S6 A0A131Z4V4 L7M5Z6 A0A131X8X4 B7QAY0 V5I2G7 A0A060WNH9 W5UMA9 A0A1S3SNS3 A0A087UQB7 A0A2C9KHX1 A0A3B1J581 A0A3Q2E4M4 A0A060WF73 B6RB61 A0A0P7ZA42 A0A0B1PKG4 A0A3P8XEZ2 A0A077Z113 K4G058 A0A232EQP0 A0A210QWI2 A0A3Q2QGL0 A0A0K8RE66 A0A3B3WP10 A0A0S7HSN9 M3ZWP3 A0A087XVF3 A0A3B3VED6 A0A3P9QC74 S4RRY0 A0A3B3RIV1 A0A3B3CFI2 K4GIG1 A0A3B3CH02 A0A1Q3EU74 Q4V9I5 A0A250Y987 A0A085MX62 A0A1B6GLV5 C9W1F3 W5N535 G3SMS7 A0A2Y9DUE0 A0A0B7AW55 A0A3Q3LQD6 A0A3Q3DKY4 E9IBD6 A0A0N0BDG3 A0A0B7AWH3 A0A224XQC8 A0A1S4EY08 T1E3B6 A0A3Q1EW71 A0A3Q1HN29 A0A3Q1BAV6 A0A3P8RK49 A0A3B4EUU4 A0A3Q2VPQ6 A0A3Q4M3M6 I3K2H9 A0A3P8UHT0 I3MQ27
PDB
6FRK
E-value=5.20483e-21,
Score=242
Ontologies
GO
Topology
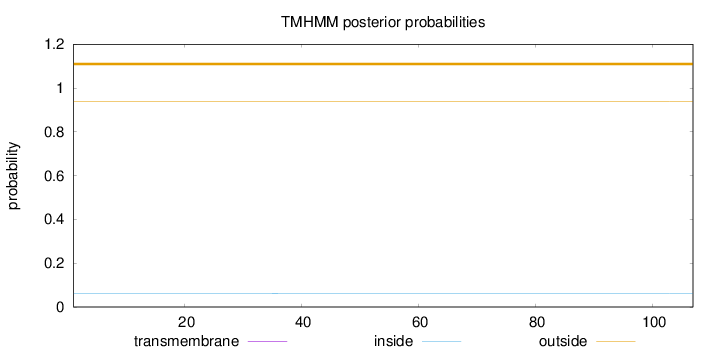
Length:
107
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000740000000000001
Exp number, first 60 AAs:
0.0004
Total prob of N-in:
0.06221
outside
1 - 107
Population Genetic Test Statistics
Pi
266.78265
Theta
206.790889
Tajima's D
0.913282
CLR
3.812116
CSRT
0.63916804159792
Interpretation
Uncertain
