Pre Gene Modal
BGIBMGA005040
Annotation
Transforming_acidic_coiled-coil-containing_protein_1_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 2.415
Sequence
CDS
ATGCAACAATTATCCGTAACACAGACTAGCAGTGGTTCTACTGAAAAGCCTACACAAGTGGTTACTCCTGCAGTTAACCGAAGTCAACCCTCCGCGAAAGTAACACCAGTAAAAGCTAGTACAGTTTCACCTGCTATAGCTACCATTGATAGATTGCTTAGTCTGGGTGCTAACATTCCTCCACCTCCACCTGTTATTACTCGTACTTCAGATATGGATCCCCATACAGTAGAACAATTGCGAGCCGTCAAAGAACTTCTTACTACTCAAGAAGAAGAAGCTCAAAGCTTGAGAGATCTTAATCATGAATTACGAGAGAGGCTTGAAGAATGTGAAAACAAAATAAAGAAATTGACTGAACAAAATGAAGAATATGCAGAGAAAGAAAAAGCCTTATCACAGAGAGTTGCAGAAAAAGTAAGAAACAATAAACAGATGAGTGTAGTTATGGAAGAATATGAGCGCACAATCTCTTCACTCATTGGTGAGCGTGAGATAGAGGGTAAAAGATGGACTGAGGAAAGAACTACACTTGTAAGAGAGAGAGATGAAGCGGCAGCACATCTTGCATCTATGGAGCATGCATTCAATGATGTTCATACCAAATATGAAAGATGTAAAGTTATCATTCAAGGCTATAAAAGCAATGAAGAGACACTCAAACATACCGTTGAGGAGAATAATGATGCCATACAAAAACTAGAAGCGAGGTATGAAACATTGAGGCAACATGCTATGCAGCAACTCAACAAAGCAAATGCTGAATTGGATGCCGTTAAAAAGACTCATCAGGCAGAAATTCTGAAATTAAATGCAATGCTCAAGAAAAGTGAGGTACATGCTTCATCACTGCAGGAGTCATTGGCACAGAAAATAAAAGACAATGAGGAACTCACAGCCATCTGTGATGAGCTCATTAATAAAGTAGGATAA
Protein
MQQLSVTQTSSGSTEKPTQVVTPAVNRSQPSAKVTPVKASTVSPAIATIDRLLSLGANIPPPPPVITRTSDMDPHTVEQLRAVKELLTTQEEEAQSLRDLNHELRERLEECENKIKKLTEQNEEYAEKEKALSQRVAEKVRNNKQMSVVMEEYERTISSLIGEREIEGKRWTEERTTLVRERDEAAAHLASMEHAFNDVHTKYERCKVIIQGYKSNEETLKHTVEENNDAIQKLEARYETLRQHAMQQLNKANAELDAVKKTHQAEILKLNAMLKKSEVHASSLQESLAQKIKDNEELTAICDELINKVG
Summary
Uniprot
H9J699
A0A2H1W7H1
A0A2A4J804
A0A2W1BQU3
A0A0L7L3B5
A0A1E1WSL9
+ More
A0A194PG72 A0A194RJW1 H9J5J7 A0A2W1BDY9 A0A2H1WST3 H9J697 A0A2A4JUQ1 A0A0L7KYT3 A0A194PGB5 A0A194RI24 A0A2P8YJD7 A0A1B6FB38 A0A212EV10 A0A1B6H2F4 A0A1B6GRL9 A0A2J7QA73 A0A1B6DLP1 A0A212FJD1 A0A2J7QA98 A0A1B6CGN1 A0A2J7QA67 S4P856 A0A1B6KP51 A0A2H1W7F0 A0A1B6E5C2 A0A1B6E6Y4 A0A1B6M8S8 A0A0L7QP40 A0A0N0BGA9 E2BM16 V9ID51 A0A026WRH9 A0A3L8DMJ0 K7J9K7 A0A2A3EQS5 E9IMI6 A0A154PKC5 E0VVR0 A0A087ZVJ2 A0A0V0G5Q5 A0A0K2UQ23 A0A0P4VJX4 A0A224XI21 A0A0K8SMS4 A0A232FBH8 A0A0A9YXA7 A0A0K8SXK2 A0A146LXJ0 A0A0A9ZGD0 A0A0K8SXY1 T1I0Y2 A0A0A9YZ01 A0A0K8SXL1 A0A0K8SXL4 A0A146LXB7 A0A210QVL7 A0A1I8MLY8 A0A1I8MLY5 A0A0L0C1C2 A0A3M6U6T7
A0A194PG72 A0A194RJW1 H9J5J7 A0A2W1BDY9 A0A2H1WST3 H9J697 A0A2A4JUQ1 A0A0L7KYT3 A0A194PGB5 A0A194RI24 A0A2P8YJD7 A0A1B6FB38 A0A212EV10 A0A1B6H2F4 A0A1B6GRL9 A0A2J7QA73 A0A1B6DLP1 A0A212FJD1 A0A2J7QA98 A0A1B6CGN1 A0A2J7QA67 S4P856 A0A1B6KP51 A0A2H1W7F0 A0A1B6E5C2 A0A1B6E6Y4 A0A1B6M8S8 A0A0L7QP40 A0A0N0BGA9 E2BM16 V9ID51 A0A026WRH9 A0A3L8DMJ0 K7J9K7 A0A2A3EQS5 E9IMI6 A0A154PKC5 E0VVR0 A0A087ZVJ2 A0A0V0G5Q5 A0A0K2UQ23 A0A0P4VJX4 A0A224XI21 A0A0K8SMS4 A0A232FBH8 A0A0A9YXA7 A0A0K8SXK2 A0A146LXJ0 A0A0A9ZGD0 A0A0K8SXY1 T1I0Y2 A0A0A9YZ01 A0A0K8SXL1 A0A0K8SXL4 A0A146LXB7 A0A210QVL7 A0A1I8MLY8 A0A1I8MLY5 A0A0L0C1C2 A0A3M6U6T7
Pubmed
EMBL
BABH01019874
ODYU01006819
SOQ49020.1
NWSH01002747
PCG67580.1
KZ149994
+ More
PZC75497.1 JTDY01003190 KOB69993.1 GDQN01001127 JAT89927.1 KQ459604 KPI92386.1 KQ460045 KPJ18118.1 BABH01025215 BABH01025216 BABH01025217 KZ150226 PZC72055.1 ODYU01010736 SOQ56036.1 NWSH01000636 PCG75122.1 JTDY01004414 KOB68189.1 KPI92068.1 KQ460205 KPJ16980.1 PYGN01000554 PSN44346.1 GECZ01022428 JAS47341.1 AGBW02012265 OWR45319.1 GECZ01000969 JAS68800.1 GECZ01004687 JAS65082.1 NEVH01016332 PNF25491.1 GEDC01025466 GEDC01010713 JAS11832.1 JAS26585.1 AGBW02008295 OWR53819.1 PNF25489.1 GEDC01024803 GEDC01023006 JAS12495.1 JAS14292.1 PNF25490.1 GAIX01004299 JAA88261.1 GEBQ01026770 JAT13207.1 SOQ49019.1 GEDC01010228 GEDC01004204 JAS27070.1 JAS33094.1 GEDC01016680 GEDC01003607 JAS20618.1 JAS33691.1 GEBQ01007642 JAT32335.1 KQ414832 KOC60393.1 KQ435792 KOX74298.1 GL449100 EFN83265.1 JR039152 JR039153 AEY58557.1 AEY58558.1 KK107128 EZA58296.1 QOIP01000006 RLU21627.1 AAZX01006880 KZ288193 PBC34143.1 GL764165 EFZ18217.1 KQ434924 KZC11680.1 DS235813 EEB17466.1 GECL01002744 JAP03380.1 HACA01022834 CDW40195.1 GDKW01003849 JAI52746.1 GFTR01008643 JAW07783.1 GBRD01011187 JAG54637.1 NNAY01000537 OXU27808.1 GBHO01006750 GBHO01006749 JAG36854.1 JAG36855.1 GBRD01007819 JAG58002.1 GDHC01006265 JAQ12364.1 GBRD01007813 JAG58008.1 GBRD01007810 JAG58011.1 ACPB03015536 GBHO01006753 JAG36851.1 GBRD01007809 JAG58012.1 GBRD01007808 JAG58013.1 GDHC01007332 JAQ11297.1 NEDP02001654 OWF52774.1 JRES01001032 KNC26057.1 RCHS01002149 RMX49337.1
PZC75497.1 JTDY01003190 KOB69993.1 GDQN01001127 JAT89927.1 KQ459604 KPI92386.1 KQ460045 KPJ18118.1 BABH01025215 BABH01025216 BABH01025217 KZ150226 PZC72055.1 ODYU01010736 SOQ56036.1 NWSH01000636 PCG75122.1 JTDY01004414 KOB68189.1 KPI92068.1 KQ460205 KPJ16980.1 PYGN01000554 PSN44346.1 GECZ01022428 JAS47341.1 AGBW02012265 OWR45319.1 GECZ01000969 JAS68800.1 GECZ01004687 JAS65082.1 NEVH01016332 PNF25491.1 GEDC01025466 GEDC01010713 JAS11832.1 JAS26585.1 AGBW02008295 OWR53819.1 PNF25489.1 GEDC01024803 GEDC01023006 JAS12495.1 JAS14292.1 PNF25490.1 GAIX01004299 JAA88261.1 GEBQ01026770 JAT13207.1 SOQ49019.1 GEDC01010228 GEDC01004204 JAS27070.1 JAS33094.1 GEDC01016680 GEDC01003607 JAS20618.1 JAS33691.1 GEBQ01007642 JAT32335.1 KQ414832 KOC60393.1 KQ435792 KOX74298.1 GL449100 EFN83265.1 JR039152 JR039153 AEY58557.1 AEY58558.1 KK107128 EZA58296.1 QOIP01000006 RLU21627.1 AAZX01006880 KZ288193 PBC34143.1 GL764165 EFZ18217.1 KQ434924 KZC11680.1 DS235813 EEB17466.1 GECL01002744 JAP03380.1 HACA01022834 CDW40195.1 GDKW01003849 JAI52746.1 GFTR01008643 JAW07783.1 GBRD01011187 JAG54637.1 NNAY01000537 OXU27808.1 GBHO01006750 GBHO01006749 JAG36854.1 JAG36855.1 GBRD01007819 JAG58002.1 GDHC01006265 JAQ12364.1 GBRD01007813 JAG58008.1 GBRD01007810 JAG58011.1 ACPB03015536 GBHO01006753 JAG36851.1 GBRD01007809 JAG58012.1 GBRD01007808 JAG58013.1 GDHC01007332 JAQ11297.1 NEDP02001654 OWF52774.1 JRES01001032 KNC26057.1 RCHS01002149 RMX49337.1
Proteomes
Interpro
SUPFAM
SSF49785
SSF49785
ProteinModelPortal
H9J699
A0A2H1W7H1
A0A2A4J804
A0A2W1BQU3
A0A0L7L3B5
A0A1E1WSL9
+ More
A0A194PG72 A0A194RJW1 H9J5J7 A0A2W1BDY9 A0A2H1WST3 H9J697 A0A2A4JUQ1 A0A0L7KYT3 A0A194PGB5 A0A194RI24 A0A2P8YJD7 A0A1B6FB38 A0A212EV10 A0A1B6H2F4 A0A1B6GRL9 A0A2J7QA73 A0A1B6DLP1 A0A212FJD1 A0A2J7QA98 A0A1B6CGN1 A0A2J7QA67 S4P856 A0A1B6KP51 A0A2H1W7F0 A0A1B6E5C2 A0A1B6E6Y4 A0A1B6M8S8 A0A0L7QP40 A0A0N0BGA9 E2BM16 V9ID51 A0A026WRH9 A0A3L8DMJ0 K7J9K7 A0A2A3EQS5 E9IMI6 A0A154PKC5 E0VVR0 A0A087ZVJ2 A0A0V0G5Q5 A0A0K2UQ23 A0A0P4VJX4 A0A224XI21 A0A0K8SMS4 A0A232FBH8 A0A0A9YXA7 A0A0K8SXK2 A0A146LXJ0 A0A0A9ZGD0 A0A0K8SXY1 T1I0Y2 A0A0A9YZ01 A0A0K8SXL1 A0A0K8SXL4 A0A146LXB7 A0A210QVL7 A0A1I8MLY8 A0A1I8MLY5 A0A0L0C1C2 A0A3M6U6T7
A0A194PG72 A0A194RJW1 H9J5J7 A0A2W1BDY9 A0A2H1WST3 H9J697 A0A2A4JUQ1 A0A0L7KYT3 A0A194PGB5 A0A194RI24 A0A2P8YJD7 A0A1B6FB38 A0A212EV10 A0A1B6H2F4 A0A1B6GRL9 A0A2J7QA73 A0A1B6DLP1 A0A212FJD1 A0A2J7QA98 A0A1B6CGN1 A0A2J7QA67 S4P856 A0A1B6KP51 A0A2H1W7F0 A0A1B6E5C2 A0A1B6E6Y4 A0A1B6M8S8 A0A0L7QP40 A0A0N0BGA9 E2BM16 V9ID51 A0A026WRH9 A0A3L8DMJ0 K7J9K7 A0A2A3EQS5 E9IMI6 A0A154PKC5 E0VVR0 A0A087ZVJ2 A0A0V0G5Q5 A0A0K2UQ23 A0A0P4VJX4 A0A224XI21 A0A0K8SMS4 A0A232FBH8 A0A0A9YXA7 A0A0K8SXK2 A0A146LXJ0 A0A0A9ZGD0 A0A0K8SXY1 T1I0Y2 A0A0A9YZ01 A0A0K8SXL1 A0A0K8SXL4 A0A146LXB7 A0A210QVL7 A0A1I8MLY8 A0A1I8MLY5 A0A0L0C1C2 A0A3M6U6T7
PDB
5NUG
E-value=0.000164296,
Score=106
Ontologies
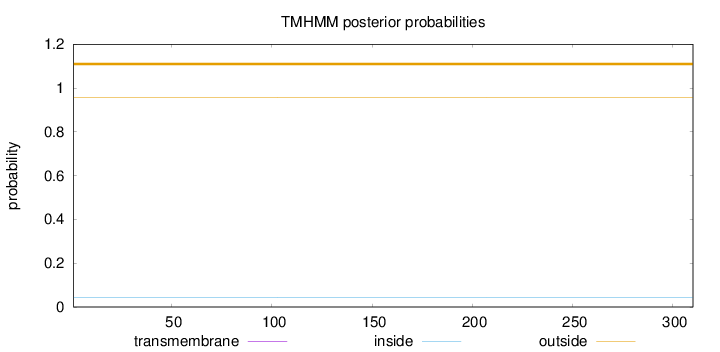
Topology
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0006
Exp number, first 60 AAs:
0.00059
Total prob of N-in:
0.04342
outside
1 - 310
Population Genetic Test Statistics
Pi
169.720086
Theta
179.48487
Tajima's D
-0.15671
CLR
0.016193
CSRT
0.327883605819709
Interpretation
Uncertain
