Pre Gene Modal
BGIBMGA005161
Annotation
glucosamine-6-phosphate_N-acetyltransferase_[Bombyx_mori]
Full name
Glucosamine 6-phosphate N-acetyltransferase
+ More
Probable glucosamine 6-phosphate N-acetyltransferase
Probable glucosamine 6-phosphate N-acetyltransferase
Alternative Name
Phosphoglucosamine acetylase
Phosphoglucosamine transacetylase
Phosphoglucosamine transacetylase
Location in the cell
Cytoplasmic Reliability : 1.749 Nuclear Reliability : 2.198
Sequence
CDS
ATGGGCATGGAAAAGAACGGAAATGCGGAAGGGAAACTCTCCGAATACCTTTACCCTCCGGAGATATTAAAAAGACTGGATTTCAGTAAGAGTCCGGCAAAGTTTCATCCTAAAATCAGTGCTGTTGACCCCGGCGAGGATTGGATGATTGTGAGGCCTTTACAACGCTCTGATTACGATAAGGGTTTCCTGCAGCTCCTCAGTCAACTAACAAGCGTGGGAAATATTACGAGAAAACAGTATGATGACCGTTTCACTAAGATGAAACACTCGGGCGGTTACTATGTGACCGTAATAGAGGACACGCGCATTAATAAACTCATAGGTGCAGCAACGCTGACCATCGAACAGAAATTCATCCACAACTGCTCTTTGCGCGGCCGTTTAGAAGACGTCGTAGTTAATGACACTTACAGAGGAAAGCAACTTGGTAAATTAATTGTGGTAACCGTATCGCTTCTCGCTCAAGAGCTGGGATGCTATAAAATGTCTCTCGACTGTAAGGATAAATTAATAAAATTTTACGAGACGCTAGGATATAAGATGGAGCCTGGGAATTCTAACGCTATGAACATGAGGTTCGATGAACCCTCTTGA
Protein
MGMEKNGNAEGKLSEYLYPPEILKRLDFSKSPAKFHPKISAVDPGEDWMIVRPLQRSDYDKGFLQLLSQLTSVGNITRKQYDDRFTKMKHSGGYYVTVIEDTRINKLIGAATLTIEQKFIHNCSLRGRLEDVVVNDTYRGKQLGKLIVVTVSLLAQELGCYKMSLDCKDKLIKFYETLGYKMEPGNSNAMNMRFDEPS
Summary
Catalytic Activity
acetyl-CoA + D-glucosamine 6-phosphate = CoA + H(+) + N-acetyl-D-glucosamine 6-phosphate
Similarity
Belongs to the acetyltransferase family. GNA1 subfamily.
Keywords
Acyltransferase
Complete proteome
Reference proteome
Transferase
Feature
chain Probable glucosamine 6-phosphate N-acetyltransferase
Uniprot
Q2F694
H9J6L9
A0A2W1BKC8
A0A2A4J851
A0A2H1W7I6
A0A194RL24
+ More
I4DRL2 A0A194PI30 S4P7A4 A0A212FJD7 A0A3S2MBB4 B4M5S9 B4JYM4 A0A0P9BMC8 A0A1B0CE03 B3MTI1 B4K610 A0A1W4UPJ6 A0A1W4UCG9 A0A1L8E462 A0A3B0K4I7 B4NBK9 A0A0B4KI57 B4PPL0 B3P5E0 Q8IMK5 B4HZD9 C6TP97 Q9VAI0 B4GPN6 A0A1I8PCI7 Q29BT7 A0A1L8E3I7 C6TP98 B4R081 A0A1I8PCU0 A0A0B4K7R2 A0A1I8PCT4 I5AMZ1 W8BP28 A0A0C9Q5S0 A0A034WIR3 A0A034WDK7 A0A0L0BN49 A0A0K8V809 A0A1I8MEF7 A0A1L8E3K6 A0A0K8TT46 A0A1I8MEF0 A0A0A1WQ62 A0A0A1WK68 A0A1A9UQ74 A0A1A9XMV1 A0A1B0BNY9 A0A1B0A8X2 D3TRP6 A0A1B0G1E0 B0WQV2 Q5Q125 Q5Q126 A0A1Q3FTP9 A0A182W600 A0A023EIL7 A0A182YH41 A0A336KA60 A0A336JZN7 A0A2M4A409 A0A182QHA9 T1DJS6 A0A182FMB2 W5JMP1 A0A2M4BZ32 A0A2M4BZ81 A0A182TRX1 A0A084VQ42 A0A182SG75 A0A2M4BZC1 A0A182V6H8 A0A182X0J7 A0A182I5T7 A0A182KV86 Q7Q212 A0A2M3ZGG6 A0A1S4H4W0 A0A182INW9 A0A182S4J3 A0A1B0D3Z6 A0A1W4X5J7 D6WTB0 A0A1J1J4W5 A0A182K3L0 A0A2P8YXE0 A0A1Y1NG47 A0A067QZM6 E0VYG6 N6TN51 A0A2L2YFS9 A0A2J7R6H8 A0A0U1U070 A0A026WXX7
I4DRL2 A0A194PI30 S4P7A4 A0A212FJD7 A0A3S2MBB4 B4M5S9 B4JYM4 A0A0P9BMC8 A0A1B0CE03 B3MTI1 B4K610 A0A1W4UPJ6 A0A1W4UCG9 A0A1L8E462 A0A3B0K4I7 B4NBK9 A0A0B4KI57 B4PPL0 B3P5E0 Q8IMK5 B4HZD9 C6TP97 Q9VAI0 B4GPN6 A0A1I8PCI7 Q29BT7 A0A1L8E3I7 C6TP98 B4R081 A0A1I8PCU0 A0A0B4K7R2 A0A1I8PCT4 I5AMZ1 W8BP28 A0A0C9Q5S0 A0A034WIR3 A0A034WDK7 A0A0L0BN49 A0A0K8V809 A0A1I8MEF7 A0A1L8E3K6 A0A0K8TT46 A0A1I8MEF0 A0A0A1WQ62 A0A0A1WK68 A0A1A9UQ74 A0A1A9XMV1 A0A1B0BNY9 A0A1B0A8X2 D3TRP6 A0A1B0G1E0 B0WQV2 Q5Q125 Q5Q126 A0A1Q3FTP9 A0A182W600 A0A023EIL7 A0A182YH41 A0A336KA60 A0A336JZN7 A0A2M4A409 A0A182QHA9 T1DJS6 A0A182FMB2 W5JMP1 A0A2M4BZ32 A0A2M4BZ81 A0A182TRX1 A0A084VQ42 A0A182SG75 A0A2M4BZC1 A0A182V6H8 A0A182X0J7 A0A182I5T7 A0A182KV86 Q7Q212 A0A2M3ZGG6 A0A1S4H4W0 A0A182INW9 A0A182S4J3 A0A1B0D3Z6 A0A1W4X5J7 D6WTB0 A0A1J1J4W5 A0A182K3L0 A0A2P8YXE0 A0A1Y1NG47 A0A067QZM6 E0VYG6 N6TN51 A0A2L2YFS9 A0A2J7R6H8 A0A0U1U070 A0A026WXX7
EC Number
2.3.1.4
Pubmed
19121390
28756777
26354079
22651552
23622113
22118469
+ More
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26109357 26109356 12537569 15632085 24495485 25348373 26108605 25315136 26369729 25830018 20353571 15857769 17510324 24945155 26483478 25244985 20920257 23761445 24438588 20966253 12364791 18362917 19820115 29403074 28004739 24845553 20566863 23537049 26561354 24508170
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26109357 26109356 12537569 15632085 24495485 25348373 26108605 25315136 26369729 25830018 20353571 15857769 17510324 24945155 26483478 25244985 20920257 23761445 24438588 20966253 12364791 18362917 19820115 29403074 28004739 24845553 20566863 23537049 26561354 24508170
EMBL
DQ311178
ABD36123.1
BABH01019875
BABH01019876
KZ149994
PZC75499.1
+ More
NWSH01002747 PCG67582.1 ODYU01006819 SOQ49018.1 KQ460045 KPJ18119.1 AK405170 BAM20552.1 KQ459604 KPI92384.1 GAIX01006471 JAA86089.1 AGBW02008295 OWR53820.1 RSAL01000001 RVE55333.1 CH940652 EDW59005.2 CH916377 EDV90786.1 CH902623 KPU72930.1 AJWK01008456 EDV30571.2 CH933806 EDW16247.2 GFDF01000772 JAV13312.1 OUUW01000007 SPP82910.1 CH964232 EDW81173.2 AE014297 AGB96461.1 CM000160 EDW98260.2 CH954182 EDV53190.1 BT132889 AAN14182.1 AEV23913.1 CH480819 EDW53396.1 BT099583 ACU43545.1 AY058687 CH479186 EDW39119.1 CM000070 EAL26910.3 GFDF01000774 JAV13310.1 BT099584 ACU43546.1 AFH06677.1 CM000364 EDX14887.1 AFH06678.1 EIM52326.2 GAMC01005918 GAMC01005917 JAC00638.1 GBYB01009443 JAG79210.1 GAKP01005294 GAKP01005292 JAC53658.1 GAKP01005296 JAC53656.1 JRES01001623 KNC21358.1 GDHF01017247 JAI35067.1 GFDF01000773 JAV13311.1 GDAI01000064 JAI17539.1 GBXI01013305 JAD00987.1 GBXI01015382 JAC98909.1 JXJN01017678 EZ424098 ADD20374.1 CCAG010015766 DS232046 EDS33032.1 AY791898 AAV84020.1 AY791896 AY791897 CH477291 AAV84018.1 AAV84019.1 EAT44583.1 GFDL01004127 JAV30918.1 JXUM01054009 JXUM01054010 JXUM01106402 GAPW01004320 KQ565051 KQ561800 JAC09278.1 KXJ71410.1 KXJ77500.1 UFQS01000035 UFQT01000035 SSW97940.1 SSX18326.1 SSW97939.1 SSX18325.1 GGFK01002164 MBW35485.1 AXCN02002602 GAMD01001296 JAB00295.1 ADMH02001125 ETN64029.1 GGFJ01009204 MBW58345.1 GGFJ01009202 MBW58343.1 ATLV01015123 KE525003 KFB40086.1 GGFJ01009203 MBW58344.1 APCN01003502 AAAB01008979 EAA13751.3 GGFM01006799 MBW27550.1 AJVK01023840 KQ971352 EFA06688.1 CVRI01000069 CRL07002.1 PYGN01000302 PSN48908.1 GEZM01003537 JAV96749.1 KK853153 KDR10580.1 DS235845 EEB18422.1 APGK01058294 KB741288 KB631652 ENN70650.1 ERL85025.1 IAAA01027634 LAA06979.1 NEVH01006756 PNF36445.1 EU252300 ACD11880.1 KK107072 EZA60653.1
NWSH01002747 PCG67582.1 ODYU01006819 SOQ49018.1 KQ460045 KPJ18119.1 AK405170 BAM20552.1 KQ459604 KPI92384.1 GAIX01006471 JAA86089.1 AGBW02008295 OWR53820.1 RSAL01000001 RVE55333.1 CH940652 EDW59005.2 CH916377 EDV90786.1 CH902623 KPU72930.1 AJWK01008456 EDV30571.2 CH933806 EDW16247.2 GFDF01000772 JAV13312.1 OUUW01000007 SPP82910.1 CH964232 EDW81173.2 AE014297 AGB96461.1 CM000160 EDW98260.2 CH954182 EDV53190.1 BT132889 AAN14182.1 AEV23913.1 CH480819 EDW53396.1 BT099583 ACU43545.1 AY058687 CH479186 EDW39119.1 CM000070 EAL26910.3 GFDF01000774 JAV13310.1 BT099584 ACU43546.1 AFH06677.1 CM000364 EDX14887.1 AFH06678.1 EIM52326.2 GAMC01005918 GAMC01005917 JAC00638.1 GBYB01009443 JAG79210.1 GAKP01005294 GAKP01005292 JAC53658.1 GAKP01005296 JAC53656.1 JRES01001623 KNC21358.1 GDHF01017247 JAI35067.1 GFDF01000773 JAV13311.1 GDAI01000064 JAI17539.1 GBXI01013305 JAD00987.1 GBXI01015382 JAC98909.1 JXJN01017678 EZ424098 ADD20374.1 CCAG010015766 DS232046 EDS33032.1 AY791898 AAV84020.1 AY791896 AY791897 CH477291 AAV84018.1 AAV84019.1 EAT44583.1 GFDL01004127 JAV30918.1 JXUM01054009 JXUM01054010 JXUM01106402 GAPW01004320 KQ565051 KQ561800 JAC09278.1 KXJ71410.1 KXJ77500.1 UFQS01000035 UFQT01000035 SSW97940.1 SSX18326.1 SSW97939.1 SSX18325.1 GGFK01002164 MBW35485.1 AXCN02002602 GAMD01001296 JAB00295.1 ADMH02001125 ETN64029.1 GGFJ01009204 MBW58345.1 GGFJ01009202 MBW58343.1 ATLV01015123 KE525003 KFB40086.1 GGFJ01009203 MBW58344.1 APCN01003502 AAAB01008979 EAA13751.3 GGFM01006799 MBW27550.1 AJVK01023840 KQ971352 EFA06688.1 CVRI01000069 CRL07002.1 PYGN01000302 PSN48908.1 GEZM01003537 JAV96749.1 KK853153 KDR10580.1 DS235845 EEB18422.1 APGK01058294 KB741288 KB631652 ENN70650.1 ERL85025.1 IAAA01027634 LAA06979.1 NEVH01006756 PNF36445.1 EU252300 ACD11880.1 KK107072 EZA60653.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000008792 UP000001070 UP000007801 UP000092461 UP000009192 UP000192221 UP000268350 UP000007798 UP000000803 UP000002282 UP000008711 UP000001292 UP000008744 UP000095300 UP000001819 UP000000304 UP000037069 UP000095301 UP000078200 UP000092443 UP000092460 UP000092445 UP000092444 UP000002320 UP000008820 UP000075920 UP000069940 UP000249989 UP000076408 UP000075886 UP000069272 UP000000673 UP000075902 UP000030765 UP000075901 UP000075903 UP000076407 UP000075840 UP000075882 UP000007062 UP000075880 UP000075900 UP000092462 UP000192223 UP000007266 UP000183832 UP000075881 UP000245037 UP000027135 UP000009046 UP000019118 UP000030742 UP000235965 UP000053097
UP000008792 UP000001070 UP000007801 UP000092461 UP000009192 UP000192221 UP000268350 UP000007798 UP000000803 UP000002282 UP000008711 UP000001292 UP000008744 UP000095300 UP000001819 UP000000304 UP000037069 UP000095301 UP000078200 UP000092443 UP000092460 UP000092445 UP000092444 UP000002320 UP000008820 UP000075920 UP000069940 UP000249989 UP000076408 UP000075886 UP000069272 UP000000673 UP000075902 UP000030765 UP000075901 UP000075903 UP000076407 UP000075840 UP000075882 UP000007062 UP000075880 UP000075900 UP000092462 UP000192223 UP000007266 UP000183832 UP000075881 UP000245037 UP000027135 UP000009046 UP000019118 UP000030742 UP000235965 UP000053097
Interpro
SUPFAM
SSF55729
SSF55729
Gene 3D
ProteinModelPortal
Q2F694
H9J6L9
A0A2W1BKC8
A0A2A4J851
A0A2H1W7I6
A0A194RL24
+ More
I4DRL2 A0A194PI30 S4P7A4 A0A212FJD7 A0A3S2MBB4 B4M5S9 B4JYM4 A0A0P9BMC8 A0A1B0CE03 B3MTI1 B4K610 A0A1W4UPJ6 A0A1W4UCG9 A0A1L8E462 A0A3B0K4I7 B4NBK9 A0A0B4KI57 B4PPL0 B3P5E0 Q8IMK5 B4HZD9 C6TP97 Q9VAI0 B4GPN6 A0A1I8PCI7 Q29BT7 A0A1L8E3I7 C6TP98 B4R081 A0A1I8PCU0 A0A0B4K7R2 A0A1I8PCT4 I5AMZ1 W8BP28 A0A0C9Q5S0 A0A034WIR3 A0A034WDK7 A0A0L0BN49 A0A0K8V809 A0A1I8MEF7 A0A1L8E3K6 A0A0K8TT46 A0A1I8MEF0 A0A0A1WQ62 A0A0A1WK68 A0A1A9UQ74 A0A1A9XMV1 A0A1B0BNY9 A0A1B0A8X2 D3TRP6 A0A1B0G1E0 B0WQV2 Q5Q125 Q5Q126 A0A1Q3FTP9 A0A182W600 A0A023EIL7 A0A182YH41 A0A336KA60 A0A336JZN7 A0A2M4A409 A0A182QHA9 T1DJS6 A0A182FMB2 W5JMP1 A0A2M4BZ32 A0A2M4BZ81 A0A182TRX1 A0A084VQ42 A0A182SG75 A0A2M4BZC1 A0A182V6H8 A0A182X0J7 A0A182I5T7 A0A182KV86 Q7Q212 A0A2M3ZGG6 A0A1S4H4W0 A0A182INW9 A0A182S4J3 A0A1B0D3Z6 A0A1W4X5J7 D6WTB0 A0A1J1J4W5 A0A182K3L0 A0A2P8YXE0 A0A1Y1NG47 A0A067QZM6 E0VYG6 N6TN51 A0A2L2YFS9 A0A2J7R6H8 A0A0U1U070 A0A026WXX7
I4DRL2 A0A194PI30 S4P7A4 A0A212FJD7 A0A3S2MBB4 B4M5S9 B4JYM4 A0A0P9BMC8 A0A1B0CE03 B3MTI1 B4K610 A0A1W4UPJ6 A0A1W4UCG9 A0A1L8E462 A0A3B0K4I7 B4NBK9 A0A0B4KI57 B4PPL0 B3P5E0 Q8IMK5 B4HZD9 C6TP97 Q9VAI0 B4GPN6 A0A1I8PCI7 Q29BT7 A0A1L8E3I7 C6TP98 B4R081 A0A1I8PCU0 A0A0B4K7R2 A0A1I8PCT4 I5AMZ1 W8BP28 A0A0C9Q5S0 A0A034WIR3 A0A034WDK7 A0A0L0BN49 A0A0K8V809 A0A1I8MEF7 A0A1L8E3K6 A0A0K8TT46 A0A1I8MEF0 A0A0A1WQ62 A0A0A1WK68 A0A1A9UQ74 A0A1A9XMV1 A0A1B0BNY9 A0A1B0A8X2 D3TRP6 A0A1B0G1E0 B0WQV2 Q5Q125 Q5Q126 A0A1Q3FTP9 A0A182W600 A0A023EIL7 A0A182YH41 A0A336KA60 A0A336JZN7 A0A2M4A409 A0A182QHA9 T1DJS6 A0A182FMB2 W5JMP1 A0A2M4BZ32 A0A2M4BZ81 A0A182TRX1 A0A084VQ42 A0A182SG75 A0A2M4BZC1 A0A182V6H8 A0A182X0J7 A0A182I5T7 A0A182KV86 Q7Q212 A0A2M3ZGG6 A0A1S4H4W0 A0A182INW9 A0A182S4J3 A0A1B0D3Z6 A0A1W4X5J7 D6WTB0 A0A1J1J4W5 A0A182K3L0 A0A2P8YXE0 A0A1Y1NG47 A0A067QZM6 E0VYG6 N6TN51 A0A2L2YFS9 A0A2J7R6H8 A0A0U1U070 A0A026WXX7
PDB
3CXS
E-value=1.00048e-37,
Score=389
Ontologies
PATHWAY
GO
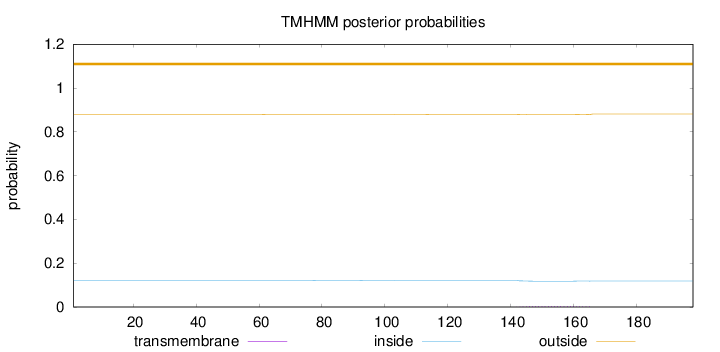
Topology
Length:
198
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08794
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.12136
outside
1 - 198
Population Genetic Test Statistics
Pi
28.416857
Theta
196.658157
Tajima's D
-1.425835
CLR
1338.576689
CSRT
0.0678966051697415
Interpretation
Uncertain
