Pre Gene Modal
BGIBMGA005037
Annotation
PREDICTED:_N-terminal_kinase-like_protein_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.357
Sequence
CDS
ATGTGGTCATTTTTCTCTCGAGATCCTACAAAGGATTTTCCGTACGATGTTGGTGAACCCGTACCAGAACTGGAAGACAGAAGTGTTTGGACATTACACAAGGGGAAAAAAAGAGGTACTCAGGATGAAGTATCAATATTTTTGTTCGACGTTCAAAAGAATTCGGAGACATTGTTTGACATAGCAAAAGCTTCTCTGAAAAGATTGAAGACTACGAGACACCCTAGCCTTTTACATTATTTAGATAGCTGTGAAACTGAGAAATTTTTGTATATAGCTACTGAATATGTAGAGCCTTTATCAACTCATTTAGAATGTATGAGGCTTGAGGGTCAGCAAAAAGACCTATTTCTTGCCTGGGGCGTATTCCAAATAACTAGAGCTTTATCATTTTTAAATAATGATGGAAATATGAGACATAACAATGTATGCCTCTATTCTGTATTTGTAACATTGGCTGGTGAATGGAAACTAGGGGGATTTGAATTTCTAACAACCCAAGGACATGAAACTACGAATCCCATACCAATAAAGATATTACCTGCCCTTGAAATATATGATCCACCAGAGAAGAATGACTCTGTAAAACTTAAAAGTGCAACTAAATGTTCATCAGACATGTGGGGACTAGGATGTCTCATATGGGAGGCATTTAATGGACCGCTCAAAAACCAACCGGCTCTTAAAACAGTAGATAATATACCAAAACAGCTATGTACGCTGTACTGTGAGCTGGTCAGTGCTAACCCTGCTTCAAGACCTAACCCGGCTGATATTATTACCAGATGTCGTAAACTTGGTGGATATTTTAAAAACGATCTGATAGATACAATGTTATTCTTAGAAGAAATTCAAATTAAAGACAAAGCTGAGAAGGGAAAATTTTTCTCAAGTCTTAGTTCACATTTAGATAACTTCCCAGAAAATGTATGTGTTCATAAGATCCTGCCGCAGTTATTGACTGCCTTTCATTATGGAGACGCTGGTTCTGCTGTGTTGGCACCTATGTTTAAGCTAGGCAAATTATTAGAAGATCCCGATTACCAAAAACAAATAGTGCCCTGTGTTGTTAAACTATTCGCCTCTAATGACAGAACGACAAGATCAAGGCTTCTGCAACAACTAGACCAGTTCATTATGCATCTGCAGAATTCGACAGTTAATGACCAAATTTTCCCGCAAGTTGTAAATGGCTTCTTGGACACAAATGCTATTATTAGGGAACAAACTGTGAAATCGATTGTTCATCTCGCTTCTAAACTAAATTACAACAATCTGAACGTTGAAGTATTGCGGCACTTTGCCAGATTACAGTCTAAAGATGATCAAGGAGGAATAAGAACGAACACAACAGTCTGCCTCGGTAAGATAGCGGCGCACCTGCATCCGCAGATACGGCAGAAGGTGCTTGTGTCTGCGTTCGTTCGCTCCACGAGGGATCCCTTCCCACCGGCGCGACAAGCCGGAGTCCTGGCGCTGGCCGCCACGCAACAGTACTTCTTGCTATCAGAAGTGGCCAACAGAGTCTTGCCGGCTCTATGTCCGCTGACAATCGATCCAGAGAAACAGGTTCGCGATGCTGCCTTTAGGACGATTAGAGGCTTTCTCGGTAAACTAGAGAAAGTTTCTGAAGACCCGAGCTTAAAAGAAGGAATGGAGGCGGACGTCCACACGGCCACTCCGTCGCTAAGCAACGCGGCGGCCACGTGGGCGGGCTGGGCCGTGACCGCCGTCACCGCCAAGTTCTACCGCTCGCACTCGGACACCACGCGGGTGCAACACTCCACTAAATCTGCACTCTCTAAACCCGGATCCTTGGAGCAACCTTCGTCGAGTAGTATGTCGACGACGACGTCATCGGTGACCTCCATGACGTCACTGGAGCACAGCGAGTCGGCCTCCGACTATGATCCGGAGCACTGGGACATGGCGGCTTGGGGAGAAATCGATTCTAGTGGAGGCACGGGCGGTAGTGCGGGCAGCAGCAGTGCGCAAGCGGCGCCCGGGCTCGCGGCCCTCGACGAGGACGACTGGGACAACGAGGAGTGGGGCACGCTGCAGGAACAACCGTCCGCCAAAGCCGATCTGGTCAGTCCCTCGGAGACGTGGAATAACTCACAATGGACGGATCCGATCGTGAACAGCGCGAACGCGACGTTCGTCCGCAGCACGGCCGGCTCGAGGCTGGAGCACCGCCCCGCCGCGCACCAGAACAGCAACGACTCGCTCGGAGGCTGGGAAGACGGAGAATTCGAACCGATAGAAGAAAACGCTGATGAAAACAATGCTTCAAAGATGGACGAGTTACGAAGGAAACGAGAAGAGAGGAAATTACAAAGACAGAGAGAGATGGAAGCGCGTCGTTCCGCTCGAGCCCAGGGACCTATGAAACTGGGCACCAAGATCGCGACAGCACCGCCGCCCTTCTAG
Protein
MWSFFSRDPTKDFPYDVGEPVPELEDRSVWTLHKGKKRGTQDEVSIFLFDVQKNSETLFDIAKASLKRLKTTRHPSLLHYLDSCETEKFLYIATEYVEPLSTHLECMRLEGQQKDLFLAWGVFQITRALSFLNNDGNMRHNNVCLYSVFVTLAGEWKLGGFEFLTTQGHETTNPIPIKILPALEIYDPPEKNDSVKLKSATKCSSDMWGLGCLIWEAFNGPLKNQPALKTVDNIPKQLCTLYCELVSANPASRPNPADIITRCRKLGGYFKNDLIDTMLFLEEIQIKDKAEKGKFFSSLSSHLDNFPENVCVHKILPQLLTAFHYGDAGSAVLAPMFKLGKLLEDPDYQKQIVPCVVKLFASNDRTTRSRLLQQLDQFIMHLQNSTVNDQIFPQVVNGFLDTNAIIREQTVKSIVHLASKLNYNNLNVEVLRHFARLQSKDDQGGIRTNTTVCLGKIAAHLHPQIRQKVLVSAFVRSTRDPFPPARQAGVLALAATQQYFLLSEVANRVLPALCPLTIDPEKQVRDAAFRTIRGFLGKLEKVSEDPSLKEGMEADVHTATPSLSNAAATWAGWAVTAVTAKFYRSHSDTTRVQHSTKSALSKPGSLEQPSSSSMSTTTSSVTSMTSLEHSESASDYDPEHWDMAAWGEIDSSGGTGGSAGSSSAQAAPGLAALDEDDWDNEEWGTLQEQPSAKADLVSPSETWNNSQWTDPIVNSANATFVRSTAGSRLEHRPAAHQNSNDSLGGWEDGEFEPIEENADENNASKMDELRRKREERKLQRQREMEARRSARAQGPMKLGTKIATAPPPF
Summary
Uniprot
H9J696
A0A2H1W7J0
A0A194RLC6
A0A194PH58
A0A3S2LWN5
A0A2A4J867
+ More
A0A067QPV9 A0A2W1BRU4 A0A2J7R6F3 A0A1W4WUG5 A0A0A1X5S0 A0A2J7R6G5 A0A0A1WGM7 A0A2J7R6G6 A0A0K8TZH6 A0A0M5J2L8 D6WR56 A0A0K8UWI1 A0A034WRN1 A0A1Y1LH31 A0A1I8NYJ2 A0A1W4WUJ0 A0A1I8N6X5 A0A0L0BN05 A0A182FJN3 A0A1S4FL31 T1PKQ5 Q16X96 B4NBL3 A0A2M3Z3I1 A0A2M4BCN2 A0A2M4BCK3 A0A1A9XMV2 A0A182Q1K3 W5JIW8 A0A2M4A403 W8C3V1 A0A1Q3FW96 A0A1Q3FW74 A0A1B0BNY8 A0A1A9W6J8 A0A1L8DNE6 A0A182RVX0 A0A1A9UQ75 A0A1L8DN65 A0A084VQ41 Q29AN9 B4G662 B4M5T3 A0A1B6DUV0 B4K614 A0A182GFH7 A0A3B0JL15 B3P5D6 B4PPK6 B3MTI5 A0A1W4URR5 Q9VAH7 B4R085 A0A1Q3G1B0 A0A1B0A8X1 B4JYM8 A0A1J1J7X2 F5HM19 Q7Q2E3 B4HZE3 U5EYR2 A0A182J9A2 A0A182YH42 A0A1B6CN54 E0VSL3 A0A336LNE4 A0A1B6LYU2 A0A1B6EK18 A0A182KGZ5 A0A182W601 A0A026WZV5 A0A151ILX4 A0A195FS56 A0A195E3C1 E9IZJ7 A0A154P2X7 A0A151I286 K7JB83 A0A158NKW6 A0A151WML9 F4WAE4 A0A088A0F8 A0A232F0K7 E2BFL8 A0A0J7NWK6 V9IAK0 A0A2A3EQZ9 A0A0L7R1K4 A0A0N1IT86 A0A2J7R6G8 E2ALL4 A0A0P5NSZ4 A0A0P5R7G3
A0A067QPV9 A0A2W1BRU4 A0A2J7R6F3 A0A1W4WUG5 A0A0A1X5S0 A0A2J7R6G5 A0A0A1WGM7 A0A2J7R6G6 A0A0K8TZH6 A0A0M5J2L8 D6WR56 A0A0K8UWI1 A0A034WRN1 A0A1Y1LH31 A0A1I8NYJ2 A0A1W4WUJ0 A0A1I8N6X5 A0A0L0BN05 A0A182FJN3 A0A1S4FL31 T1PKQ5 Q16X96 B4NBL3 A0A2M3Z3I1 A0A2M4BCN2 A0A2M4BCK3 A0A1A9XMV2 A0A182Q1K3 W5JIW8 A0A2M4A403 W8C3V1 A0A1Q3FW96 A0A1Q3FW74 A0A1B0BNY8 A0A1A9W6J8 A0A1L8DNE6 A0A182RVX0 A0A1A9UQ75 A0A1L8DN65 A0A084VQ41 Q29AN9 B4G662 B4M5T3 A0A1B6DUV0 B4K614 A0A182GFH7 A0A3B0JL15 B3P5D6 B4PPK6 B3MTI5 A0A1W4URR5 Q9VAH7 B4R085 A0A1Q3G1B0 A0A1B0A8X1 B4JYM8 A0A1J1J7X2 F5HM19 Q7Q2E3 B4HZE3 U5EYR2 A0A182J9A2 A0A182YH42 A0A1B6CN54 E0VSL3 A0A336LNE4 A0A1B6LYU2 A0A1B6EK18 A0A182KGZ5 A0A182W601 A0A026WZV5 A0A151ILX4 A0A195FS56 A0A195E3C1 E9IZJ7 A0A154P2X7 A0A151I286 K7JB83 A0A158NKW6 A0A151WML9 F4WAE4 A0A088A0F8 A0A232F0K7 E2BFL8 A0A0J7NWK6 V9IAK0 A0A2A3EQZ9 A0A0L7R1K4 A0A0N1IT86 A0A2J7R6G8 E2ALL4 A0A0P5NSZ4 A0A0P5R7G3
Pubmed
19121390
26354079
24845553
28756777
25830018
18362917
+ More
19820115 25348373 28004739 25315136 26108605 17510324 17994087 20920257 23761445 24495485 24438588 15632085 18057021 26483478 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 25244985 20566863 24508170 30249741 21282665 20075255 21347285 21719571 28648823 20798317
19820115 25348373 28004739 25315136 26108605 17510324 17994087 20920257 23761445 24495485 24438588 15632085 18057021 26483478 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 25244985 20566863 24508170 30249741 21282665 20075255 21347285 21719571 28648823 20798317
EMBL
BABH01019876
BABH01019877
ODYU01006819
SOQ49017.1
KQ460045
KPJ18120.1
+ More
KQ459604 KPI92383.1 RSAL01000001 RVE55334.1 NWSH01002747 PCG67583.1 KK853153 KDR10579.1 KZ149994 PZC75500.1 NEVH01006756 PNF36417.1 GBXI01008207 JAD06085.1 PNF36420.1 GBXI01016637 JAC97654.1 PNF36419.1 GDHF01032467 JAI19847.1 CP012526 ALC48061.1 KQ971354 EFA06008.2 GDHF01021282 JAI31032.1 GAKP01002524 JAC56428.1 GEZM01055791 JAV72934.1 JRES01001623 KNC21367.1 KA648695 AFP63324.1 CH477545 EAT39222.1 CH964232 EDW81177.2 GGFM01002321 MBW23072.1 GGFJ01001636 MBW50777.1 GGFJ01001635 MBW50776.1 AXCN02002602 ADMH02001048 ETN64322.1 GGFK01002212 MBW35533.1 GAMC01005889 JAC00667.1 GFDL01003176 JAV31869.1 GFDL01003174 JAV31871.1 JXJN01017678 GFDF01006224 JAV07860.1 GFDF01006223 JAV07861.1 ATLV01015123 KE525003 KFB40085.1 CM000070 EAL27310.3 CH479179 EDW23815.1 CH940652 EDW59009.2 KRF78641.1 GEDC01007856 JAS29442.1 CH933806 EDW16251.2 KRG02027.1 JXUM01059614 JXUM01059615 JXUM01059616 KQ562062 KXJ76769.1 OUUW01000007 SPP82915.1 CH954182 EDV53186.1 CM000160 EDW98256.1 CH902623 EDV30575.2 AE014297 BT032863 AAF56933.1 ACD81877.1 AGB96462.1 CM000364 EDX14891.1 GFDL01001462 JAV33583.1 CH916377 EDV90790.1 CVRI01000069 CRL07001.1 AAAB01008976 EGK97330.1 EAA13516.5 CH480819 EDW53400.1 GANO01000123 JAB59748.1 GEDC01022454 JAS14844.1 DS235751 EEB16369.1 UFQT01000020 SSX17897.1 GEBQ01011187 JAT28790.1 GECZ01031489 JAS38280.1 KK107072 QOIP01000007 EZA60654.1 RLU20871.1 KQ977085 KYN05891.1 KQ981281 KYN43293.1 KQ979701 KYN19660.1 GL767232 EFZ14003.1 KQ434809 KZC06182.1 KQ976551 KYM80873.1 ADTU01018978 KQ982937 KYQ49106.1 GL888048 EGI68827.1 NNAY01001403 OXU24089.1 GL448037 EFN85540.1 LBMM01001164 KMQ96800.1 JR037246 AEY57662.1 KZ288194 PBC33924.1 KQ414667 KOC64723.1 KQ435863 KOX70378.1 PNF36416.1 GL440609 EFN65623.1 GDIQ01159669 JAK92056.1 GDIQ01106529 JAL45197.1
KQ459604 KPI92383.1 RSAL01000001 RVE55334.1 NWSH01002747 PCG67583.1 KK853153 KDR10579.1 KZ149994 PZC75500.1 NEVH01006756 PNF36417.1 GBXI01008207 JAD06085.1 PNF36420.1 GBXI01016637 JAC97654.1 PNF36419.1 GDHF01032467 JAI19847.1 CP012526 ALC48061.1 KQ971354 EFA06008.2 GDHF01021282 JAI31032.1 GAKP01002524 JAC56428.1 GEZM01055791 JAV72934.1 JRES01001623 KNC21367.1 KA648695 AFP63324.1 CH477545 EAT39222.1 CH964232 EDW81177.2 GGFM01002321 MBW23072.1 GGFJ01001636 MBW50777.1 GGFJ01001635 MBW50776.1 AXCN02002602 ADMH02001048 ETN64322.1 GGFK01002212 MBW35533.1 GAMC01005889 JAC00667.1 GFDL01003176 JAV31869.1 GFDL01003174 JAV31871.1 JXJN01017678 GFDF01006224 JAV07860.1 GFDF01006223 JAV07861.1 ATLV01015123 KE525003 KFB40085.1 CM000070 EAL27310.3 CH479179 EDW23815.1 CH940652 EDW59009.2 KRF78641.1 GEDC01007856 JAS29442.1 CH933806 EDW16251.2 KRG02027.1 JXUM01059614 JXUM01059615 JXUM01059616 KQ562062 KXJ76769.1 OUUW01000007 SPP82915.1 CH954182 EDV53186.1 CM000160 EDW98256.1 CH902623 EDV30575.2 AE014297 BT032863 AAF56933.1 ACD81877.1 AGB96462.1 CM000364 EDX14891.1 GFDL01001462 JAV33583.1 CH916377 EDV90790.1 CVRI01000069 CRL07001.1 AAAB01008976 EGK97330.1 EAA13516.5 CH480819 EDW53400.1 GANO01000123 JAB59748.1 GEDC01022454 JAS14844.1 DS235751 EEB16369.1 UFQT01000020 SSX17897.1 GEBQ01011187 JAT28790.1 GECZ01031489 JAS38280.1 KK107072 QOIP01000007 EZA60654.1 RLU20871.1 KQ977085 KYN05891.1 KQ981281 KYN43293.1 KQ979701 KYN19660.1 GL767232 EFZ14003.1 KQ434809 KZC06182.1 KQ976551 KYM80873.1 ADTU01018978 KQ982937 KYQ49106.1 GL888048 EGI68827.1 NNAY01001403 OXU24089.1 GL448037 EFN85540.1 LBMM01001164 KMQ96800.1 JR037246 AEY57662.1 KZ288194 PBC33924.1 KQ414667 KOC64723.1 KQ435863 KOX70378.1 PNF36416.1 GL440609 EFN65623.1 GDIQ01159669 JAK92056.1 GDIQ01106529 JAL45197.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000218220
UP000027135
+ More
UP000235965 UP000192223 UP000092553 UP000007266 UP000095300 UP000095301 UP000037069 UP000069272 UP000008820 UP000007798 UP000092443 UP000075886 UP000000673 UP000092460 UP000091820 UP000075900 UP000078200 UP000030765 UP000001819 UP000008744 UP000008792 UP000009192 UP000069940 UP000249989 UP000268350 UP000008711 UP000002282 UP000007801 UP000192221 UP000000803 UP000000304 UP000092445 UP000001070 UP000183832 UP000007062 UP000001292 UP000075880 UP000076408 UP000009046 UP000075881 UP000075920 UP000053097 UP000279307 UP000078542 UP000078541 UP000078492 UP000076502 UP000078540 UP000002358 UP000005205 UP000075809 UP000007755 UP000005203 UP000215335 UP000008237 UP000036403 UP000242457 UP000053825 UP000053105 UP000000311
UP000235965 UP000192223 UP000092553 UP000007266 UP000095300 UP000095301 UP000037069 UP000069272 UP000008820 UP000007798 UP000092443 UP000075886 UP000000673 UP000092460 UP000091820 UP000075900 UP000078200 UP000030765 UP000001819 UP000008744 UP000008792 UP000009192 UP000069940 UP000249989 UP000268350 UP000008711 UP000002282 UP000007801 UP000192221 UP000000803 UP000000304 UP000092445 UP000001070 UP000183832 UP000007062 UP000001292 UP000075880 UP000076408 UP000009046 UP000075881 UP000075920 UP000053097 UP000279307 UP000078542 UP000078541 UP000078492 UP000076502 UP000078540 UP000002358 UP000005205 UP000075809 UP000007755 UP000005203 UP000215335 UP000008237 UP000036403 UP000242457 UP000053825 UP000053105 UP000000311
Interpro
Gene 3D
ProteinModelPortal
H9J696
A0A2H1W7J0
A0A194RLC6
A0A194PH58
A0A3S2LWN5
A0A2A4J867
+ More
A0A067QPV9 A0A2W1BRU4 A0A2J7R6F3 A0A1W4WUG5 A0A0A1X5S0 A0A2J7R6G5 A0A0A1WGM7 A0A2J7R6G6 A0A0K8TZH6 A0A0M5J2L8 D6WR56 A0A0K8UWI1 A0A034WRN1 A0A1Y1LH31 A0A1I8NYJ2 A0A1W4WUJ0 A0A1I8N6X5 A0A0L0BN05 A0A182FJN3 A0A1S4FL31 T1PKQ5 Q16X96 B4NBL3 A0A2M3Z3I1 A0A2M4BCN2 A0A2M4BCK3 A0A1A9XMV2 A0A182Q1K3 W5JIW8 A0A2M4A403 W8C3V1 A0A1Q3FW96 A0A1Q3FW74 A0A1B0BNY8 A0A1A9W6J8 A0A1L8DNE6 A0A182RVX0 A0A1A9UQ75 A0A1L8DN65 A0A084VQ41 Q29AN9 B4G662 B4M5T3 A0A1B6DUV0 B4K614 A0A182GFH7 A0A3B0JL15 B3P5D6 B4PPK6 B3MTI5 A0A1W4URR5 Q9VAH7 B4R085 A0A1Q3G1B0 A0A1B0A8X1 B4JYM8 A0A1J1J7X2 F5HM19 Q7Q2E3 B4HZE3 U5EYR2 A0A182J9A2 A0A182YH42 A0A1B6CN54 E0VSL3 A0A336LNE4 A0A1B6LYU2 A0A1B6EK18 A0A182KGZ5 A0A182W601 A0A026WZV5 A0A151ILX4 A0A195FS56 A0A195E3C1 E9IZJ7 A0A154P2X7 A0A151I286 K7JB83 A0A158NKW6 A0A151WML9 F4WAE4 A0A088A0F8 A0A232F0K7 E2BFL8 A0A0J7NWK6 V9IAK0 A0A2A3EQZ9 A0A0L7R1K4 A0A0N1IT86 A0A2J7R6G8 E2ALL4 A0A0P5NSZ4 A0A0P5R7G3
A0A067QPV9 A0A2W1BRU4 A0A2J7R6F3 A0A1W4WUG5 A0A0A1X5S0 A0A2J7R6G5 A0A0A1WGM7 A0A2J7R6G6 A0A0K8TZH6 A0A0M5J2L8 D6WR56 A0A0K8UWI1 A0A034WRN1 A0A1Y1LH31 A0A1I8NYJ2 A0A1W4WUJ0 A0A1I8N6X5 A0A0L0BN05 A0A182FJN3 A0A1S4FL31 T1PKQ5 Q16X96 B4NBL3 A0A2M3Z3I1 A0A2M4BCN2 A0A2M4BCK3 A0A1A9XMV2 A0A182Q1K3 W5JIW8 A0A2M4A403 W8C3V1 A0A1Q3FW96 A0A1Q3FW74 A0A1B0BNY8 A0A1A9W6J8 A0A1L8DNE6 A0A182RVX0 A0A1A9UQ75 A0A1L8DN65 A0A084VQ41 Q29AN9 B4G662 B4M5T3 A0A1B6DUV0 B4K614 A0A182GFH7 A0A3B0JL15 B3P5D6 B4PPK6 B3MTI5 A0A1W4URR5 Q9VAH7 B4R085 A0A1Q3G1B0 A0A1B0A8X1 B4JYM8 A0A1J1J7X2 F5HM19 Q7Q2E3 B4HZE3 U5EYR2 A0A182J9A2 A0A182YH42 A0A1B6CN54 E0VSL3 A0A336LNE4 A0A1B6LYU2 A0A1B6EK18 A0A182KGZ5 A0A182W601 A0A026WZV5 A0A151ILX4 A0A195FS56 A0A195E3C1 E9IZJ7 A0A154P2X7 A0A151I286 K7JB83 A0A158NKW6 A0A151WML9 F4WAE4 A0A088A0F8 A0A232F0K7 E2BFL8 A0A0J7NWK6 V9IAK0 A0A2A3EQZ9 A0A0L7R1K4 A0A0N1IT86 A0A2J7R6G8 E2ALL4 A0A0P5NSZ4 A0A0P5R7G3
PDB
3VWA
E-value=3.46116e-20,
Score=245
Ontologies
GO
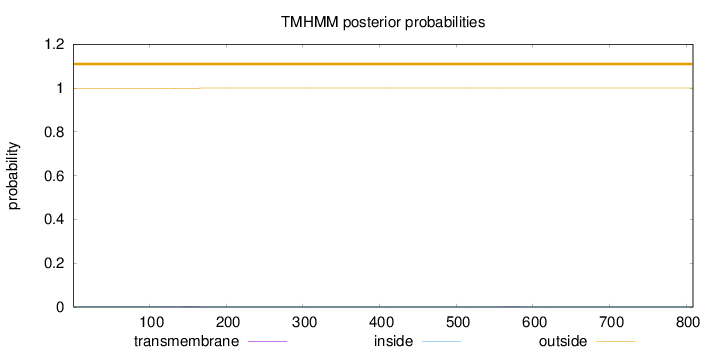
Topology
Length:
809
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0457900000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00171
outside
1 - 809
Population Genetic Test Statistics
Pi
203.865907
Theta
179.010084
Tajima's D
0.394242
CLR
0.636677
CSRT
0.486175691215439
Interpretation
Possibly Positive selection
