Gene
KWMTBOMO15265
Pre Gene Modal
BGIBMGA005029
Annotation
PREDICTED:_nephrin-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.279 PlasmaMembrane Reliability : 1.339
Sequence
CDS
ATGAATTACGACATAGTATTACGATTGTCTACGGTCGATGTGGATGCAGTCCTAGGGCGCACTGCGTCCTTGCCCTGTGACGTCACGCCCGACGCCAACGAAGACCGCGTGTACATGGTGCTCTGGTTCAGGAAAAGTGGAGGGAAACCTATATACAGCTTTGACGTACGGGGGCGTTCATTCAACAAAGCTCTACAATGGTCGGACCCTGGCGCTTTCGGACCACGAGCTTACTTCGCAACTGTCGCTCGACCGGCGGCCCTGACCTTGACCTCCGTGCAACTTGACGACGAAGGCGTATACCGATGCAGAGTAGACTTCAAGAATTCACCCACAAGAAATTTTCAAATACGGCTGACTGTTATCGTGCCTCCTCATCAAATGTTATTATATGACAAGTCAGGAGCTGATGTGTCTGGAGTCGTAGGACCTTTGGAAGAAGGTGCAGCATTAGTTCTGATCTGCGAAGTACGCGGAGGTCGTCCAGAGCCGCTTGTTTCTTGGTGGATAAACGGCGTACCGGCTCCTCAATACATTGGTATCAAAACCGATACCCACGTGGTGGTGAACAGGCTCGAACACCAAAATTTGAAACGAGATGATCTGAACACGACCTTCAAATGTAGAGCTACAAATACTAATTTGGTTCCACCCCAAGAGAAAGTTGTAAGGCTGGAGTTGAATCTGTCTCCGCTTTCCGTTTCTTTGCTGAACCGACCGCAGCAGTTGTTAGCCGGCTCAGCTGCGACCATACGATGCGTATCCGAAGGCAGCCGACCTCCAGCACAGATCTCATGGTTTAAAGACAACAGGAGTTTTTCAAAAAACATGATATCGGATCACTCAAATGAAACGTGGTCACTCAGTGCGTTAGTGTTCACTCCAGCGCCTACGGATCACGGAACTACAATCAAATGCGTCGCAGCGAACCCTAGCCTACCTACTAAGACTATTAACGATCAGCTACTGCTTAATGTTGTATATAGTCCCCTGGTGATCCTGACTTTGGGCAGCACTCTCAACCCAGACGACATCAAAGAAGGGGACGACGTTTACTTCGAGTGCAATGTCCGAGCGAATCCTCGGGAGCATCGAATATCATGGTACCACAATGACAGGCCGGTGATGCAGAATATGTCTTCGGGTATCATCGTGAGTACGCGGTCTCTGGTTCTGCAGAAGGTTACGAGAGGAGAAGCCGGCGCCTATACTTGCAAAGCGGCTAATTCTCGAGGCGAAACTTCTAGTCAAGCTGTGAAATTGAGAGTGCAATATGCTCCGGTTTGCTCTGAACCATCTCCTCAAGTAGTAGGGGCCGCAATAGATGAGGCTTTACAAGTTCGATGCGCGGTCCACGCGGACCCTTCAGACGTAACTTTCTTATGGCAATTCAACAATAGCGGAGAGAGTTTCGACGTTTCTCCAGCTAGATATGGTACAATCAACAGCAGTGTAAGTGAGCTTCGGTACAGGCCATCAAGTGAAAGGGATTATGGAGCTCTAACATGCAGTGGTATTAACACAGTAGGTCGGCAAACACAGCCCTGCGTTTTCCAAATTGTTCCAGCTGCTCGCCCCTCACCTCCAAAGAATTGCACAGTAACGTCTAGTAACAATTCGACTTGGCTGGAAGACTCCAAAGAAGAAACGGAAGATTTTCTGATTGTGCGATGTGTGACCGGCTATGACGGCGGTCTTCCACAACTGGTTGTACTGGAAGCAGTTGATTCTGACAGTGGAACTGTTAGATTCAATGTAACTGCCAATGAAACAGATGGAGTTGCCTTCTTCTCGGTGCCTTCAGGAGTTTTGTGGTCGAGTAGAAATGCTTTAAGACTCACCGTACATTCTAGGAACGATAAAGGATCCTCTGAACGTATTTTGCTTCAAGCACTCACTTACCAAGGACCGGAAAGGAGAACAGATGGAGTCAATAACTTAGTAGCCGGAGTATCTAAAACTGCGTTTTGGGCCATAATTGTTACATCTGTGATCACCGCCAGCGCACTGATACTCATCGTGTATTTTTTGAGAAGAAAGAGGGAGAGACCGACAATGAAACAATCATCAGAGCTTCAAATCAACGCTGGATCCGGTCAGTATGTGGTAGCGTACACACTAAAACCTTCTAAACAAGCTCAACCTGATATCTTAAATCCACCTTCTGATGGGGGCACTCAAATTGCTAAGGGCGTTGCAGGGACTGTCAAAAATGGCACTCTACCCATGAGTGACGAGTGGTCCGCCGGCGAAAGAGCCCAGTACGCGTCGTCGCCGAAAACGTCACTCTCAAGTAAGCCAGCTGCCTCTGGGACCACCACGCTCAGTAGGAGAGAGCATCTCATAGCTGAAGAGATCCCAGGACCGGAGAGTTGCGTATAA
Protein
MNYDIVLRLSTVDVDAVLGRTASLPCDVTPDANEDRVYMVLWFRKSGGKPIYSFDVRGRSFNKALQWSDPGAFGPRAYFATVARPAALTLTSVQLDDEGVYRCRVDFKNSPTRNFQIRLTVIVPPHQMLLYDKSGADVSGVVGPLEEGAALVLICEVRGGRPEPLVSWWINGVPAPQYIGIKTDTHVVVNRLEHQNLKRDDLNTTFKCRATNTNLVPPQEKVVRLELNLSPLSVSLLNRPQQLLAGSAATIRCVSEGSRPPAQISWFKDNRSFSKNMISDHSNETWSLSALVFTPAPTDHGTTIKCVAANPSLPTKTINDQLLLNVVYSPLVILTLGSTLNPDDIKEGDDVYFECNVRANPREHRISWYHNDRPVMQNMSSGIIVSTRSLVLQKVTRGEAGAYTCKAANSRGETSSQAVKLRVQYAPVCSEPSPQVVGAAIDEALQVRCAVHADPSDVTFLWQFNNSGESFDVSPARYGTINSSVSELRYRPSSERDYGALTCSGINTVGRQTQPCVFQIVPAARPSPPKNCTVTSSNNSTWLEDSKEETEDFLIVRCVTGYDGGLPQLVVLEAVDSDSGTVRFNVTANETDGVAFFSVPSGVLWSSRNALRLTVHSRNDKGSSERILLQALTYQGPERRTDGVNNLVAGVSKTAFWAIIVTSVITASALILIVYFLRRKRERPTMKQSSELQINAGSGQYVVAYTLKPSKQAQPDILNPPSDGGTQIAKGVAGTVKNGTLPMSDEWSAGERAQYASSPKTSLSSKPAASGTTTLSRREHLIAEEIPGPESCV
Summary
Uniprot
A0A3S2P2C6
A0A2H1V4B0
A0A194RLD1
A0A212FKA1
A0A194PGE6
A0A2A4JIS9
+ More
A0A194RL28 A0A2W1BQV1 A0A0L7LLH3 A0A212F6U3 A0A3S2L969 A0A1W4WMZ2 A0A1W4WBP5 A0A154PH73 T1HZW4 A0A3L8D739 E2BN54 A0A2A3ES63 A0A087ZZQ1 A0A195BHH6 A0A026WJB3 A0A195EGH0 F4WG39 A0A195C8J0 A0A195F0N4 E2AKZ8 A0A151XBQ1 A0A158NT21 A0A232F405 K7IQE1 Q0KIB1 A0A2A4JZZ9 A0A2W1BE05 A0A0L7R1L0 A0A0C9RJZ2 A0A0M8ZVF6 A0A1W4WLT4 A0A1W4WMR9 D6WPK9 A0A1B6G259 A0A0K8T359 A0A0A9ZBH5 A0A1B6LB71 A0A336M9R3 A0A067RCL4 A0A0L0BY62 A0A1I8PBW5 A0A1Y1NKS5 A0A1B6C6V9 A0A0Q9W1D8 B4JI81 B4M6A4 A0A139WE49 A0A0M5JCQ5 B4KAA3 B3LX91 A0A1I8NIL5 A0A2H1WJ89 A0A1W4WRT9 A0A1W4WQT7 A0A1W4WFY8 A0A1W4UR97 A0A1W4V3H8 A0A2J7PYN3 A0A034W9N8 C0PTV4 A0A1W4V371 A0A182IS29 B4NKE4 W8B9C8 A0A0R3NN42 A0A3B0KV58 B4G2W0 Q297Q6
A0A194RL28 A0A2W1BQV1 A0A0L7LLH3 A0A212F6U3 A0A3S2L969 A0A1W4WMZ2 A0A1W4WBP5 A0A154PH73 T1HZW4 A0A3L8D739 E2BN54 A0A2A3ES63 A0A087ZZQ1 A0A195BHH6 A0A026WJB3 A0A195EGH0 F4WG39 A0A195C8J0 A0A195F0N4 E2AKZ8 A0A151XBQ1 A0A158NT21 A0A232F405 K7IQE1 Q0KIB1 A0A2A4JZZ9 A0A2W1BE05 A0A0L7R1L0 A0A0C9RJZ2 A0A0M8ZVF6 A0A1W4WLT4 A0A1W4WMR9 D6WPK9 A0A1B6G259 A0A0K8T359 A0A0A9ZBH5 A0A1B6LB71 A0A336M9R3 A0A067RCL4 A0A0L0BY62 A0A1I8PBW5 A0A1Y1NKS5 A0A1B6C6V9 A0A0Q9W1D8 B4JI81 B4M6A4 A0A139WE49 A0A0M5JCQ5 B4KAA3 B3LX91 A0A1I8NIL5 A0A2H1WJ89 A0A1W4WRT9 A0A1W4WQT7 A0A1W4WFY8 A0A1W4UR97 A0A1W4V3H8 A0A2J7PYN3 A0A034W9N8 C0PTV4 A0A1W4V371 A0A182IS29 B4NKE4 W8B9C8 A0A0R3NN42 A0A3B0KV58 B4G2W0 Q297Q6
Pubmed
26354079
22118469
28756777
26227816
30249741
20798317
+ More
24508170 21719571 21347285 28648823 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18362917 19820115 25401762 26823975 24845553 26108605 28004739 17994087 18057021 25315136 25348373 24495485 15632085 23185243
24508170 21719571 21347285 28648823 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18362917 19820115 25401762 26823975 24845553 26108605 28004739 17994087 18057021 25315136 25348373 24495485 15632085 23185243
EMBL
RSAL01000001
RVE55340.1
ODYU01000599
SOQ35673.1
KQ460045
KPJ18125.1
+ More
AGBW02008103 OWR54162.1 KQ459604 KPI92377.1 NWSH01001251 PCG71977.1 KPJ18124.1 KZ149994 PZC75507.1 JTDY01000670 KOB76275.1 AGBW02009972 OWR49444.1 RSAL01000082 RVE48510.1 KQ434905 KZC11236.1 ACPB03001712 QOIP01000012 RLU15688.1 GL449385 EFN82816.1 KZ288194 PBC33881.1 KQ976488 KYM83604.1 KK107185 EZA55761.1 KQ978957 KYN26989.1 GL888128 EGI66806.1 KQ978205 KYM96491.1 KQ981897 KYN33674.1 GL440425 EFN65915.1 KQ982320 KYQ57806.1 ADTU01025477 ADTU01025478 ADTU01025479 ADTU01025480 NNAY01001084 OXU25200.1 AE014297 ABI31143.1 NWSH01000305 PCG77591.1 KZ150231 PZC71994.1 KQ414667 KOC64681.1 GBYB01013606 GBYB01013607 JAG83373.1 JAG83374.1 KQ435863 KOX70423.1 KQ971354 EFA06826.2 GECZ01013250 JAS56519.1 GBRD01006221 JAG59600.1 GBHO01002348 GDHC01008300 JAG41256.1 JAQ10329.1 GEBQ01019020 JAT20957.1 UFQT01000721 SSX26760.1 KK852551 KDR21492.1 JRES01001160 KNC24967.1 GEZM01002471 JAV97235.1 GEDC01028050 JAS09248.1 CH940652 KRF78738.1 CH916369 EDV92962.1 EDW59180.1 KRF78739.1 KRF78740.1 KYB26230.1 CP012526 ALC47181.1 CH933806 EDW14590.1 KRG01093.1 KRG01094.1 CH902617 EDV41691.1 KPU79296.1 KPU79297.1 KPU79299.1 ODYU01008707 SOQ52514.1 NEVH01020344 PNF21422.1 GAKP01006691 JAC52261.1 BT071799 ACN43735.2 CH964272 EDW84074.1 GAMC01011313 JAB95242.1 CM000070 KRT00496.1 OUUW01000013 SPP87858.1 CH479179 EDW24155.1 EAL28149.2 KRT00494.1 KRT00495.1 KRT00497.1
AGBW02008103 OWR54162.1 KQ459604 KPI92377.1 NWSH01001251 PCG71977.1 KPJ18124.1 KZ149994 PZC75507.1 JTDY01000670 KOB76275.1 AGBW02009972 OWR49444.1 RSAL01000082 RVE48510.1 KQ434905 KZC11236.1 ACPB03001712 QOIP01000012 RLU15688.1 GL449385 EFN82816.1 KZ288194 PBC33881.1 KQ976488 KYM83604.1 KK107185 EZA55761.1 KQ978957 KYN26989.1 GL888128 EGI66806.1 KQ978205 KYM96491.1 KQ981897 KYN33674.1 GL440425 EFN65915.1 KQ982320 KYQ57806.1 ADTU01025477 ADTU01025478 ADTU01025479 ADTU01025480 NNAY01001084 OXU25200.1 AE014297 ABI31143.1 NWSH01000305 PCG77591.1 KZ150231 PZC71994.1 KQ414667 KOC64681.1 GBYB01013606 GBYB01013607 JAG83373.1 JAG83374.1 KQ435863 KOX70423.1 KQ971354 EFA06826.2 GECZ01013250 JAS56519.1 GBRD01006221 JAG59600.1 GBHO01002348 GDHC01008300 JAG41256.1 JAQ10329.1 GEBQ01019020 JAT20957.1 UFQT01000721 SSX26760.1 KK852551 KDR21492.1 JRES01001160 KNC24967.1 GEZM01002471 JAV97235.1 GEDC01028050 JAS09248.1 CH940652 KRF78738.1 CH916369 EDV92962.1 EDW59180.1 KRF78739.1 KRF78740.1 KYB26230.1 CP012526 ALC47181.1 CH933806 EDW14590.1 KRG01093.1 KRG01094.1 CH902617 EDV41691.1 KPU79296.1 KPU79297.1 KPU79299.1 ODYU01008707 SOQ52514.1 NEVH01020344 PNF21422.1 GAKP01006691 JAC52261.1 BT071799 ACN43735.2 CH964272 EDW84074.1 GAMC01011313 JAB95242.1 CM000070 KRT00496.1 OUUW01000013 SPP87858.1 CH479179 EDW24155.1 EAL28149.2 KRT00494.1 KRT00495.1 KRT00497.1
Proteomes
UP000283053
UP000053240
UP000007151
UP000053268
UP000218220
UP000037510
+ More
UP000192223 UP000076502 UP000015103 UP000279307 UP000008237 UP000242457 UP000005203 UP000078540 UP000053097 UP000078492 UP000007755 UP000078542 UP000078541 UP000000311 UP000075809 UP000005205 UP000215335 UP000002358 UP000000803 UP000053825 UP000053105 UP000007266 UP000027135 UP000037069 UP000095300 UP000008792 UP000001070 UP000092553 UP000009192 UP000007801 UP000095301 UP000192221 UP000235965 UP000075880 UP000007798 UP000001819 UP000268350 UP000008744
UP000192223 UP000076502 UP000015103 UP000279307 UP000008237 UP000242457 UP000005203 UP000078540 UP000053097 UP000078492 UP000007755 UP000078542 UP000078541 UP000000311 UP000075809 UP000005205 UP000215335 UP000002358 UP000000803 UP000053825 UP000053105 UP000007266 UP000027135 UP000037069 UP000095300 UP000008792 UP000001070 UP000092553 UP000009192 UP000007801 UP000095301 UP000192221 UP000235965 UP000075880 UP000007798 UP000001819 UP000268350 UP000008744
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A3S2P2C6
A0A2H1V4B0
A0A194RLD1
A0A212FKA1
A0A194PGE6
A0A2A4JIS9
+ More
A0A194RL28 A0A2W1BQV1 A0A0L7LLH3 A0A212F6U3 A0A3S2L969 A0A1W4WMZ2 A0A1W4WBP5 A0A154PH73 T1HZW4 A0A3L8D739 E2BN54 A0A2A3ES63 A0A087ZZQ1 A0A195BHH6 A0A026WJB3 A0A195EGH0 F4WG39 A0A195C8J0 A0A195F0N4 E2AKZ8 A0A151XBQ1 A0A158NT21 A0A232F405 K7IQE1 Q0KIB1 A0A2A4JZZ9 A0A2W1BE05 A0A0L7R1L0 A0A0C9RJZ2 A0A0M8ZVF6 A0A1W4WLT4 A0A1W4WMR9 D6WPK9 A0A1B6G259 A0A0K8T359 A0A0A9ZBH5 A0A1B6LB71 A0A336M9R3 A0A067RCL4 A0A0L0BY62 A0A1I8PBW5 A0A1Y1NKS5 A0A1B6C6V9 A0A0Q9W1D8 B4JI81 B4M6A4 A0A139WE49 A0A0M5JCQ5 B4KAA3 B3LX91 A0A1I8NIL5 A0A2H1WJ89 A0A1W4WRT9 A0A1W4WQT7 A0A1W4WFY8 A0A1W4UR97 A0A1W4V3H8 A0A2J7PYN3 A0A034W9N8 C0PTV4 A0A1W4V371 A0A182IS29 B4NKE4 W8B9C8 A0A0R3NN42 A0A3B0KV58 B4G2W0 Q297Q6
A0A194RL28 A0A2W1BQV1 A0A0L7LLH3 A0A212F6U3 A0A3S2L969 A0A1W4WMZ2 A0A1W4WBP5 A0A154PH73 T1HZW4 A0A3L8D739 E2BN54 A0A2A3ES63 A0A087ZZQ1 A0A195BHH6 A0A026WJB3 A0A195EGH0 F4WG39 A0A195C8J0 A0A195F0N4 E2AKZ8 A0A151XBQ1 A0A158NT21 A0A232F405 K7IQE1 Q0KIB1 A0A2A4JZZ9 A0A2W1BE05 A0A0L7R1L0 A0A0C9RJZ2 A0A0M8ZVF6 A0A1W4WLT4 A0A1W4WMR9 D6WPK9 A0A1B6G259 A0A0K8T359 A0A0A9ZBH5 A0A1B6LB71 A0A336M9R3 A0A067RCL4 A0A0L0BY62 A0A1I8PBW5 A0A1Y1NKS5 A0A1B6C6V9 A0A0Q9W1D8 B4JI81 B4M6A4 A0A139WE49 A0A0M5JCQ5 B4KAA3 B3LX91 A0A1I8NIL5 A0A2H1WJ89 A0A1W4WRT9 A0A1W4WQT7 A0A1W4WFY8 A0A1W4UR97 A0A1W4V3H8 A0A2J7PYN3 A0A034W9N8 C0PTV4 A0A1W4V371 A0A182IS29 B4NKE4 W8B9C8 A0A0R3NN42 A0A3B0KV58 B4G2W0 Q297Q6
PDB
5ZO2
E-value=8.20237e-14,
Score=190
Ontologies
GO
Topology
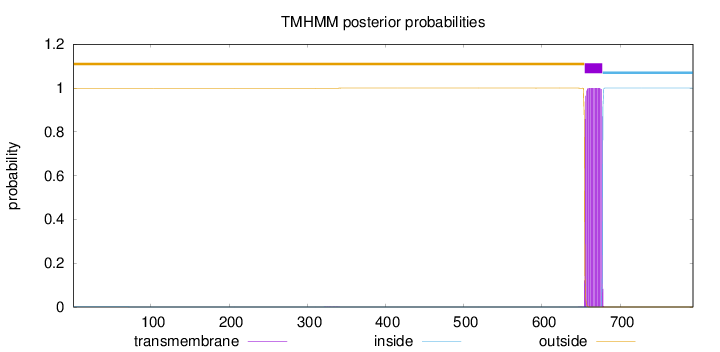
Length:
793
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.89428
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.00060
outside
1 - 654
TMhelix
655 - 677
inside
678 - 793
Population Genetic Test Statistics
Pi
246.134084
Theta
172.33702
Tajima's D
1.427316
CLR
0.525052
CSRT
0.777511124443778
Interpretation
Uncertain
