Gene
KWMTBOMO15263
Pre Gene Modal
BGIBMGA009757
Annotation
PREDICTED:_mucin-2-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.679
Sequence
CDS
ATGAAACACATTCCTAAAGAAAAAAGACAGAAATGGGACAAAAAGTCTGAACAATGCATACTTATAGGCTATCCTGAAGATGTTAAGGGATACCGTATATACAATCCTGCAACACAGAGCATCACAACAAGTCGAGATGTTATATTTATAGAAGAAAATAACTTAGATGAACAGAAAATCATTGAAATAAAAGAAGATGATTTAAGTAATAAATCATTGTCACCTTCAGTGGGAGATTTACCGGACACCAGTAGGGAGGATTCATTTACTTCAGCTGAAAATGCTACTGACTTGGATGAAACCTACGTACCATCTTCTGAGGATGAGGAGTCCTTGAAATTAGAAAAAGACCTGCTGAGAACATTACCGAAAAGGCACAGAAAGCAACCTGACAGATATGGATATATGTGTACAGAAGAATGTAACAATCCAGTCAGAGATGAATTGACATTAGAAGAGGCACTGGAAGGACCAGAAAAGGAGCACTGGGAACAGGCTATCAAAGAAGAACTGAAAAGTTTTGAAGACAACAAGGCATGGGATATTGTTGACGTTCCTGTAAGTGGCAACATTGTAAAGTGCAAGTGGGTTCTTAAAAAGAAATACGATGCTGAAAATAATGTGCGATATCGAGCGAGACTTGTGGCTAAAGGTTGTTCACAAAGATATGGTGAGGACTACAGTGAAACTTTCTCTCCTGTGGTGCGGCATACAACACTTAGATTATTATTTGCTTTATCAGTGCAGATGAACCTGAAAGTGATTCATCTTGATGTAAAAACTGCCTTCTTGAATGGAGATCTGTCTGAGACTATATACATGCAAAAACCTACAGGCTATGTCTGTAAAGATGATAAAAAGGTTCTTAAATTAAACAAAGCTATATATGGTCTCAAGCAGGCTTCACGAGCATGGTATATGAAGGTTGATGCTTTAAAACTATTGTAA
Protein
MKHIPKEKRQKWDKKSEQCILIGYPEDVKGYRIYNPATQSITTSRDVIFIEENNLDEQKIIEIKEDDLSNKSLSPSVGDLPDTSREDSFTSAENATDLDETYVPSSEDEESLKLEKDLLRTLPKRHRKQPDRYGYMCTEECNNPVRDELTLEEALEGPEKEHWEQAIKEELKSFEDNKAWDIVDVPVSGNIVKCKWVLKKKYDAENNVRYRARLVAKGCSQRYGEDYSETFSPVVRHTTLRLLFALSVQMNLKVIHLDVKTAFLNGDLSETIYMQKPTGYVCKDDKKVLKLNKAIYGLKQASRAWYMKVDALKLL
Summary
Uniprot
A0A1Y1NAZ7
A0A1Y1N668
A0A1Y1KL90
A0A212F5G1
A0A1Y1N664
A0A194RJV9
+ More
A0A1Y1N932 A0A0N1ICY1 A0A0A9XV68 A0A0A9XY84 A0A0A9WQD4 A0A146LH62 A0A182H8F8 T1PKV2 A0A0C9RAA9 A0A355BBC5 A0A0J7NHJ8 A0A034VP70 A0A2A4JRI1 A0A0A9XUP7 Q9ZRJ0 A0A2N9H8E1 A0A0J7K416 A0A2N9H4V1 A0A2N9IT68 A0A2N9GRB2 A0A0J7K6A2 A0A2N9I2Y6 A0A2N9HJ76 A0A2N9GYW9 A0A2N9GMY7 A0A2N9I6R9 A0A2N9H7K3 A0A2N9ELY1 A0A2N9G4M2 A0A2N9EXI3 A0A2N9I8F7 A0A2N9GX47 A0A2N9EMG6 A0A2N9FSJ8 A0A2N9IWT1 A0A2N9FV67 A0A2N9EM80 A0A2N9GA80 A0A1Y1LYU4 A0A2N9EHW3 A0A2N9GGS7 A0A2N9FRN5 A0A2N9FW82 A0A2N9ETC8 A0A2N9FL83 A0A2N9EYI8 A0A2N9EIR6 A0A2N9GHK3 A0A2N9FVE8 A0A2N9HZR4 A0A2N9EK90 A0A2N9GMG5 A0A2N9I7K4 A0A2N9FQJ1 A0A2N9FHB7 A0A392MBJ0 A0A2N9FWL6 A0A2N9G940 A0A2N9EGT6 A0A396GW39 A0A0A9YB53 A0A0A9W6P7 A0A2N9GQ16 A0A2N9JB46 A0A2N9HKJ4 A0A2N9IGQ9 A0A2N9GCG9 A0A2N9HZ75 A0A2N9HJF5 A0A2N9I1P0 A0A2N9GRZ0 A0A2N9I9F7 A0A2N9IGN3 A0A2A4JA71 A0A2N9GEX0 A0A0J7K6W1 A0A2K3NXK0 A0A2N9H6B2 A0A2N9I6N4 A0A2N9J6C2 A0A2N9IN51 A0A2N9EI16 A0A2N9I0Q5 A0A1W7R6H7 A0A2N9G9X7 A0A151TS30 A0A0J7K6U3 K7JC54 A0A2N9GBF3 W8C3B0 A0A2N9H0T6
A0A1Y1N932 A0A0N1ICY1 A0A0A9XV68 A0A0A9XY84 A0A0A9WQD4 A0A146LH62 A0A182H8F8 T1PKV2 A0A0C9RAA9 A0A355BBC5 A0A0J7NHJ8 A0A034VP70 A0A2A4JRI1 A0A0A9XUP7 Q9ZRJ0 A0A2N9H8E1 A0A0J7K416 A0A2N9H4V1 A0A2N9IT68 A0A2N9GRB2 A0A0J7K6A2 A0A2N9I2Y6 A0A2N9HJ76 A0A2N9GYW9 A0A2N9GMY7 A0A2N9I6R9 A0A2N9H7K3 A0A2N9ELY1 A0A2N9G4M2 A0A2N9EXI3 A0A2N9I8F7 A0A2N9GX47 A0A2N9EMG6 A0A2N9FSJ8 A0A2N9IWT1 A0A2N9FV67 A0A2N9EM80 A0A2N9GA80 A0A1Y1LYU4 A0A2N9EHW3 A0A2N9GGS7 A0A2N9FRN5 A0A2N9FW82 A0A2N9ETC8 A0A2N9FL83 A0A2N9EYI8 A0A2N9EIR6 A0A2N9GHK3 A0A2N9FVE8 A0A2N9HZR4 A0A2N9EK90 A0A2N9GMG5 A0A2N9I7K4 A0A2N9FQJ1 A0A2N9FHB7 A0A392MBJ0 A0A2N9FWL6 A0A2N9G940 A0A2N9EGT6 A0A396GW39 A0A0A9YB53 A0A0A9W6P7 A0A2N9GQ16 A0A2N9JB46 A0A2N9HKJ4 A0A2N9IGQ9 A0A2N9GCG9 A0A2N9HZ75 A0A2N9HJF5 A0A2N9I1P0 A0A2N9GRZ0 A0A2N9I9F7 A0A2N9IGN3 A0A2A4JA71 A0A2N9GEX0 A0A0J7K6W1 A0A2K3NXK0 A0A2N9H6B2 A0A2N9I6N4 A0A2N9J6C2 A0A2N9IN51 A0A2N9EI16 A0A2N9I0Q5 A0A1W7R6H7 A0A2N9G9X7 A0A151TS30 A0A0J7K6U3 K7JC54 A0A2N9GBF3 W8C3B0 A0A2N9H0T6
Pubmed
EMBL
GEZM01011855
JAV93396.1
GEZM01011856
JAV93393.1
GEZM01082909
JAV61231.1
+ More
AGBW02010203 OWR48976.1 GEZM01011859 JAV93384.1 KQ460297 KPJ16236.1 GEZM01011857 JAV93390.1 KQ460882 KPJ11482.1 GBHO01019780 JAG23824.1 GBHO01019781 JAG23823.1 GBHO01036544 JAG07060.1 GDHC01011760 JAQ06869.1 JXUM01118474 KQ566190 KXJ70335.1 KA648745 AFP63374.1 GBYB01005195 JAG74962.1 DNYX01000169 HBK83011.1 LBMM01004924 KMQ91985.1 GAKP01015352 JAC43600.1 NWSH01000816 PCG74073.1 GBHO01019985 JAG23619.1 D83003 BAA11674.1 OIVN01002984 SPD07941.1 LBMM01014433 KMQ85183.1 OIVN01002854 SPD06945.1 OIVN01006189 SPD27343.1 OIVN01002268 SPD02135.1 LBMM01013184 KMQ85774.1 OIVN01004614 SPD18379.1 OIVN01003897 SPD14257.1 OIVN01002564 SPD04723.1 OIVN01002137 SPD00935.1 OIVN01004921 SPD19975.1 OIVN01002940 SPD07631.1 OIVN01000183 SPC75833.1 OIVN01001484 SPC94512.1 OIVN01000395 SPC79545.1 OIVN01005068 SPD20708.1 OIVN01002491 SPD04083.1 OIVN01000191 SPC75975.1 OIVN01001130 SPC90232.1 OIVN01006238 SPD28613.1 OIVN01001196 SPC91020.1 OIVN01000189 SPC75922.1 OIVN01001666 SPC96375.1 GEZM01045163 GEZM01045162 JAV77888.1 OIVN01000095 SPC74169.1 OIVN01001890 SPC98658.1 OIVN01001091 SPC89740.1 OIVN01001520 SPC94877.1 OIVN01000557 SPC82046.1 OIVN01000958 SPC87933.1 OIVN01000404 SPC79669.1 OIVN01000116 SPC74628.1 OIVN01002247 SPD01987.1 OIVN01001202 SPC91100.1 OIVN01004420 SPD17230.1 OIVN01000147 SPC75222.1 OIVN01002444 SPD03657.1 OIVN01005334 SPD21977.1 OIVN01001052 SPC89229.1 OIVN01000866 SPC86622.1 LXQA010007518 MCH84877.1 OIVN01001534 SPC95017.1 OIVN01001636 SPC96102.1 OIVN01000080 SPC73890.1 PSQE01000007 RHN45312.1 GBHO01016859 JAG26745.1 GBHO01043059 JAG00545.1 OIVN01002547 SPD04547.1 OIVN01006468 SPD33689.1 OIVN01003569 SPD12194.1 OIVN01005646 SPD23424.1 OIVN01001733 SPC97079.1 OIVN01004379 SPD17029.1 OIVN01003558 SPD12118.1 OIVN01004603 SPD18338.1 OIVN01002256 SPD02034.1 OIVN01005059 SPD20680.1 OIVN01005591 SPD23209.1 NWSH01002292 PCG68656.1 OIVN01001789 SPC97674.1 LBMM01012645 KMQ86082.1 ASHM01002084 PNY07765.1 OIVN01002904 SPD07378.1 OIVN01004879 SPD19723.1 OIVN01006429 SPD32977.1 OIVN01006120 SPD25513.1 OIVN01000106 SPC74385.1 OIVN01004867 SPD19656.1 GEHC01000879 JAV46766.1 OIVN01001656 SPC96275.1 CM003605 KYP69858.1 LBMM01012492 KMQ86193.1 AAZX01015857 AAZX01019482 OIVN01001669 SPC96464.1 GAMC01006169 JAC00387.1 OIVN01002680 SPD05612.1
AGBW02010203 OWR48976.1 GEZM01011859 JAV93384.1 KQ460297 KPJ16236.1 GEZM01011857 JAV93390.1 KQ460882 KPJ11482.1 GBHO01019780 JAG23824.1 GBHO01019781 JAG23823.1 GBHO01036544 JAG07060.1 GDHC01011760 JAQ06869.1 JXUM01118474 KQ566190 KXJ70335.1 KA648745 AFP63374.1 GBYB01005195 JAG74962.1 DNYX01000169 HBK83011.1 LBMM01004924 KMQ91985.1 GAKP01015352 JAC43600.1 NWSH01000816 PCG74073.1 GBHO01019985 JAG23619.1 D83003 BAA11674.1 OIVN01002984 SPD07941.1 LBMM01014433 KMQ85183.1 OIVN01002854 SPD06945.1 OIVN01006189 SPD27343.1 OIVN01002268 SPD02135.1 LBMM01013184 KMQ85774.1 OIVN01004614 SPD18379.1 OIVN01003897 SPD14257.1 OIVN01002564 SPD04723.1 OIVN01002137 SPD00935.1 OIVN01004921 SPD19975.1 OIVN01002940 SPD07631.1 OIVN01000183 SPC75833.1 OIVN01001484 SPC94512.1 OIVN01000395 SPC79545.1 OIVN01005068 SPD20708.1 OIVN01002491 SPD04083.1 OIVN01000191 SPC75975.1 OIVN01001130 SPC90232.1 OIVN01006238 SPD28613.1 OIVN01001196 SPC91020.1 OIVN01000189 SPC75922.1 OIVN01001666 SPC96375.1 GEZM01045163 GEZM01045162 JAV77888.1 OIVN01000095 SPC74169.1 OIVN01001890 SPC98658.1 OIVN01001091 SPC89740.1 OIVN01001520 SPC94877.1 OIVN01000557 SPC82046.1 OIVN01000958 SPC87933.1 OIVN01000404 SPC79669.1 OIVN01000116 SPC74628.1 OIVN01002247 SPD01987.1 OIVN01001202 SPC91100.1 OIVN01004420 SPD17230.1 OIVN01000147 SPC75222.1 OIVN01002444 SPD03657.1 OIVN01005334 SPD21977.1 OIVN01001052 SPC89229.1 OIVN01000866 SPC86622.1 LXQA010007518 MCH84877.1 OIVN01001534 SPC95017.1 OIVN01001636 SPC96102.1 OIVN01000080 SPC73890.1 PSQE01000007 RHN45312.1 GBHO01016859 JAG26745.1 GBHO01043059 JAG00545.1 OIVN01002547 SPD04547.1 OIVN01006468 SPD33689.1 OIVN01003569 SPD12194.1 OIVN01005646 SPD23424.1 OIVN01001733 SPC97079.1 OIVN01004379 SPD17029.1 OIVN01003558 SPD12118.1 OIVN01004603 SPD18338.1 OIVN01002256 SPD02034.1 OIVN01005059 SPD20680.1 OIVN01005591 SPD23209.1 NWSH01002292 PCG68656.1 OIVN01001789 SPC97674.1 LBMM01012645 KMQ86082.1 ASHM01002084 PNY07765.1 OIVN01002904 SPD07378.1 OIVN01004879 SPD19723.1 OIVN01006429 SPD32977.1 OIVN01006120 SPD25513.1 OIVN01000106 SPC74385.1 OIVN01004867 SPD19656.1 GEHC01000879 JAV46766.1 OIVN01001656 SPC96275.1 CM003605 KYP69858.1 LBMM01012492 KMQ86193.1 AAZX01015857 AAZX01019482 OIVN01001669 SPC96464.1 GAMC01006169 JAC00387.1 OIVN01002680 SPD05612.1
Proteomes
Pfam
Interpro
IPR025724
GAG-pre-integrase_dom
+ More
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR013103 RVT_2
IPR001584 Integrase_cat-core
IPR039537 Retrotran_Ty1/copia-like
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR025314 DUF4219
IPR032675 LRR_dom_sf
IPR036576 WRKY_dom_sf
IPR001611 Leu-rich_rpt
IPR003657 WRKY_dom
IPR002182 NB-ARC
IPR027417 P-loop_NTPase
IPR041118 Rx_N
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR013103 RVT_2
IPR001584 Integrase_cat-core
IPR039537 Retrotran_Ty1/copia-like
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR025314 DUF4219
IPR032675 LRR_dom_sf
IPR036576 WRKY_dom_sf
IPR001611 Leu-rich_rpt
IPR003657 WRKY_dom
IPR002182 NB-ARC
IPR027417 P-loop_NTPase
IPR041118 Rx_N
Gene 3D
ProteinModelPortal
A0A1Y1NAZ7
A0A1Y1N668
A0A1Y1KL90
A0A212F5G1
A0A1Y1N664
A0A194RJV9
+ More
A0A1Y1N932 A0A0N1ICY1 A0A0A9XV68 A0A0A9XY84 A0A0A9WQD4 A0A146LH62 A0A182H8F8 T1PKV2 A0A0C9RAA9 A0A355BBC5 A0A0J7NHJ8 A0A034VP70 A0A2A4JRI1 A0A0A9XUP7 Q9ZRJ0 A0A2N9H8E1 A0A0J7K416 A0A2N9H4V1 A0A2N9IT68 A0A2N9GRB2 A0A0J7K6A2 A0A2N9I2Y6 A0A2N9HJ76 A0A2N9GYW9 A0A2N9GMY7 A0A2N9I6R9 A0A2N9H7K3 A0A2N9ELY1 A0A2N9G4M2 A0A2N9EXI3 A0A2N9I8F7 A0A2N9GX47 A0A2N9EMG6 A0A2N9FSJ8 A0A2N9IWT1 A0A2N9FV67 A0A2N9EM80 A0A2N9GA80 A0A1Y1LYU4 A0A2N9EHW3 A0A2N9GGS7 A0A2N9FRN5 A0A2N9FW82 A0A2N9ETC8 A0A2N9FL83 A0A2N9EYI8 A0A2N9EIR6 A0A2N9GHK3 A0A2N9FVE8 A0A2N9HZR4 A0A2N9EK90 A0A2N9GMG5 A0A2N9I7K4 A0A2N9FQJ1 A0A2N9FHB7 A0A392MBJ0 A0A2N9FWL6 A0A2N9G940 A0A2N9EGT6 A0A396GW39 A0A0A9YB53 A0A0A9W6P7 A0A2N9GQ16 A0A2N9JB46 A0A2N9HKJ4 A0A2N9IGQ9 A0A2N9GCG9 A0A2N9HZ75 A0A2N9HJF5 A0A2N9I1P0 A0A2N9GRZ0 A0A2N9I9F7 A0A2N9IGN3 A0A2A4JA71 A0A2N9GEX0 A0A0J7K6W1 A0A2K3NXK0 A0A2N9H6B2 A0A2N9I6N4 A0A2N9J6C2 A0A2N9IN51 A0A2N9EI16 A0A2N9I0Q5 A0A1W7R6H7 A0A2N9G9X7 A0A151TS30 A0A0J7K6U3 K7JC54 A0A2N9GBF3 W8C3B0 A0A2N9H0T6
A0A1Y1N932 A0A0N1ICY1 A0A0A9XV68 A0A0A9XY84 A0A0A9WQD4 A0A146LH62 A0A182H8F8 T1PKV2 A0A0C9RAA9 A0A355BBC5 A0A0J7NHJ8 A0A034VP70 A0A2A4JRI1 A0A0A9XUP7 Q9ZRJ0 A0A2N9H8E1 A0A0J7K416 A0A2N9H4V1 A0A2N9IT68 A0A2N9GRB2 A0A0J7K6A2 A0A2N9I2Y6 A0A2N9HJ76 A0A2N9GYW9 A0A2N9GMY7 A0A2N9I6R9 A0A2N9H7K3 A0A2N9ELY1 A0A2N9G4M2 A0A2N9EXI3 A0A2N9I8F7 A0A2N9GX47 A0A2N9EMG6 A0A2N9FSJ8 A0A2N9IWT1 A0A2N9FV67 A0A2N9EM80 A0A2N9GA80 A0A1Y1LYU4 A0A2N9EHW3 A0A2N9GGS7 A0A2N9FRN5 A0A2N9FW82 A0A2N9ETC8 A0A2N9FL83 A0A2N9EYI8 A0A2N9EIR6 A0A2N9GHK3 A0A2N9FVE8 A0A2N9HZR4 A0A2N9EK90 A0A2N9GMG5 A0A2N9I7K4 A0A2N9FQJ1 A0A2N9FHB7 A0A392MBJ0 A0A2N9FWL6 A0A2N9G940 A0A2N9EGT6 A0A396GW39 A0A0A9YB53 A0A0A9W6P7 A0A2N9GQ16 A0A2N9JB46 A0A2N9HKJ4 A0A2N9IGQ9 A0A2N9GCG9 A0A2N9HZ75 A0A2N9HJF5 A0A2N9I1P0 A0A2N9GRZ0 A0A2N9I9F7 A0A2N9IGN3 A0A2A4JA71 A0A2N9GEX0 A0A0J7K6W1 A0A2K3NXK0 A0A2N9H6B2 A0A2N9I6N4 A0A2N9J6C2 A0A2N9IN51 A0A2N9EI16 A0A2N9I0Q5 A0A1W7R6H7 A0A2N9G9X7 A0A151TS30 A0A0J7K6U3 K7JC54 A0A2N9GBF3 W8C3B0 A0A2N9H0T6
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Nucleus
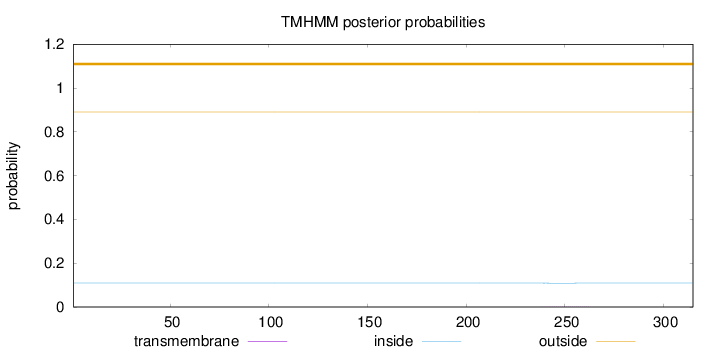
Length:
315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0113
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10945
outside
1 - 315
Population Genetic Test Statistics
Pi
4.490763
Theta
30.839539
Tajima's D
-0.377636
CLR
4.830238
CSRT
0.269986500674966
Interpretation
Uncertain
