Gene
KWMTBOMO15262 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005025
Annotation
PREDICTED:_uncharacterized_protein_LOC101743760_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.64
Sequence
CDS
ATGAAAGTCGCTATTGTGTTTATGGGCTTGATCGCTCTTGGCTTCTCGGTGCCCCGACCGAAGAAGAGCTTCGTAGAAAACTTTGGTGATTTCCTTGACATCATTAAGGACGAGGCTGGTAATGATATCGATAATTTGTTCGAGCAATACATCGAATTTGAGGAGTTCCGGCGTTCGCTTGACTACCTCACCACGAAGGACTTCAGAGACTTGATCTATGAGATGGAAGACCTGCCGGAATTCAAAGCTGTGGTCGATTTCCTGGAAAATGACAACATTGAGATCCATTTCTTCATTGATATTTTCAACGAAATGATGGAAACTATCGGCGAGAGAGTTAAAAGAGCCCGACAAACTCTTAGCGGACGCGACTTCACTTCTTACATTAACGATATTATCGATGTATTCCCCAAGGATAAGCTCGCAGCCTTGTACGAACAGAAACTCGCTGAGGACGAAGAATTCAGAGTAGCTCTTGAGAACCTCCAGAGCGAAGAATGGGACGCCGTGTTCGGTGCTCTGTGGGAGAGTGAGCAATTCAAGGCCGAAGTTGATACTCTTGCTGAACACGGTATTGACATACACGTCCTCATGAAAGAGCTCTTCGCCATCTTCGGACAAAACTAA
Protein
MKVAIVFMGLIALGFSVPRPKKSFVENFGDFLDIIKDEAGNDIDNLFEQYIEFEEFRRSLDYLTTKDFRDLIYEMEDLPEFKAVVDFLENDNIEIHFFIDIFNEMMETIGERVKRARQTLSGRDFTSYINDIIDVFPKDKLAALYEQKLAEDEEFRVALENLQSEEWDAVFGALWESEQFKAEVDTLAEHGIDIHVLMKELFAIFGQN
Summary
Uniprot
H9J684
H9J683
B2KSG9
B2KSG6
B2KSI3
A0A2H1W637
+ More
B2KSH0 A0A2W1BRV7 B2KSH1 A0A161H9I2 A0A194RQ23 B2KSI5 A0A194PI21 A0A2A4J0T9 B2KSI4 B2KXE0 A0A0L7LQ68 B2KSG5 B2KSG7 B2KSG8 B2KSH6 B2KSH4 B2KSG4 B2KXE2 A0A212EQZ3 B2KSI2 B2KXD9 B2KSH7 A0A3S2LJN8 B2KSG3 B2KSH9 B2KSI1 B2KSH8 B2KXD8 B2KSH5 B2KSH2 B2KXD7 Q6TAW0 B2KSH3 A0A026WXX3 A0A026WX70 B2KSI0 A0A067QVM1 A0A3L8E2R5 E9J3M2 A0A0C9PIB4 A0A2P8Y6L1 E2A6C9 A0A0L7R895 A0A2P8YVU1 Q16K34 A0A151XFV9 A8C9U5 A0A151JU60 A0A195CV20 Q16WN9 A0A2A3E602 A9XFX0 A0A182G0S0 O96522 A0A088AIT4 U5EVT0 Q16K33 A0A1L8DRE9 A0A1L8DR00 F4MI46 A0A087ZXS1 A0A2P0XIZ6 Q16WN7 A0A2J7R8A2 Q9TZR6 O18535 U5EMJ5 A0A182G0S2 K7J3S2 A0A158NFC6 A0A195E8S7 O18530 D3JUE8 Q16K36 A0A0N0BIK3 B0X1R6 A0A195BI59 A0A026WXQ5 A0A1L8DR35 F4X027 A0A2J7R8A9 E2A6E2 A0A1L8DR95 O18527 A0A182G6A6 A8CW22 A0A182H7M5 A0A087ZXS3 O18528 F4MI43 A0A023ELN6 A0A195E9N4 D4NTK1
B2KSH0 A0A2W1BRV7 B2KSH1 A0A161H9I2 A0A194RQ23 B2KSI5 A0A194PI21 A0A2A4J0T9 B2KSI4 B2KXE0 A0A0L7LQ68 B2KSG5 B2KSG7 B2KSG8 B2KSH6 B2KSH4 B2KSG4 B2KXE2 A0A212EQZ3 B2KSI2 B2KXD9 B2KSH7 A0A3S2LJN8 B2KSG3 B2KSH9 B2KSI1 B2KSH8 B2KXD8 B2KSH5 B2KSH2 B2KXD7 Q6TAW0 B2KSH3 A0A026WXX3 A0A026WX70 B2KSI0 A0A067QVM1 A0A3L8E2R5 E9J3M2 A0A0C9PIB4 A0A2P8Y6L1 E2A6C9 A0A0L7R895 A0A2P8YVU1 Q16K34 A0A151XFV9 A8C9U5 A0A151JU60 A0A195CV20 Q16WN9 A0A2A3E602 A9XFX0 A0A182G0S0 O96522 A0A088AIT4 U5EVT0 Q16K33 A0A1L8DRE9 A0A1L8DR00 F4MI46 A0A087ZXS1 A0A2P0XIZ6 Q16WN7 A0A2J7R8A2 Q9TZR6 O18535 U5EMJ5 A0A182G0S2 K7J3S2 A0A158NFC6 A0A195E8S7 O18530 D3JUE8 Q16K36 A0A0N0BIK3 B0X1R6 A0A195BI59 A0A026WXQ5 A0A1L8DR35 F4X027 A0A2J7R8A9 E2A6E2 A0A1L8DR95 O18527 A0A182G6A6 A8CW22 A0A182H7M5 A0A087ZXS3 O18528 F4MI43 A0A023ELN6 A0A195E9N4 D4NTK1
Pubmed
EMBL
BABH01019931
EU137123
ABX39543.1
EU137120
EU265821
ABX39540.1
+ More
EU137137 ABX39557.1 ODYU01006273 SOQ47964.1 EU137124 ABX39544.1 KZ150046 PZC74493.1 EU137125 ABX39545.1 KT984952 AMY63171.1 KQ460045 KPJ18126.1 EU137139 ABX39559.1 KQ459604 KPI92374.1 NWSH01003917 PCG65694.1 PCG65695.1 EU137138 ABX39558.1 EU265820 ABY88947.1 JTDY01000350 KOB77590.1 EU137119 ABX39539.1 EU137121 ABX39541.1 EU137122 ABX39542.1 EU137130 ABX39550.1 EU137128 ABX39548.1 EU137118 ABX39538.1 EU265822 ABY88949.1 AGBW02013167 OWR43916.1 EU137136 ABX39556.1 EU265819 ABY88946.1 EU137131 ABX39551.1 RSAL01000001 RVE55341.1 EU137117 ABX39537.1 EU137133 ABX39553.1 EU137135 ABX39555.1 EU137132 ABX39552.1 EU265818 ABY88945.1 EU137129 ABX39549.1 EU137126 ABX39546.1 EU265817 ABY88944.1 AY425622 AAR84202.1 EU137127 ABX39547.1 KK107077 EZA60591.1 QOIP01000001 EZA60607.1 RLU26326.1 RLU26700.1 EU137134 ABX39554.1 KK853323 KDR08524.1 RLU27030.1 GL768066 EFZ12622.1 GBYB01000663 JAG70430.1 PYGN01000874 PSN39804.1 GL437123 EFN71014.1 KQ414633 KOC67100.1 PYGN01000332 PSN48368.1 CH477978 CH477559 EAT34668.1 EAT38996.1 KQ982182 KYQ59195.1 EU031911 ABV44704.1 KQ981791 KYN35782.1 KQ977279 KYN04372.1 EAT38997.1 KZ288364 PBC26914.1 EF202179 ABP04045.1 JXUM01006083 KQ560208 KXJ83828.1 AF072220 AAD13531.1 GANO01001740 JAB58131.1 EAT34669.1 GFDF01005179 JAV08905.1 GFDF01005178 JAV08906.1 EZ933294 ADJ57675.1 KY661332 AVA17419.1 EAT38998.1 NEVH01006723 PNF37061.1 AF072222 AAD13533.1 U78970 AAC34312.1 GANO01000999 JAB58872.1 JXUM01006095 KXJ83830.1 ADTU01014245 KQ979440 KYN21516.1 U69957 AAB82404.1 GU301883 ADB92492.1 EAT34666.1 KQ435729 KOX77672.1 DS232267 EDS38794.1 KQ976464 KYM84538.1 EZA60593.1 GFDF01005185 JAV08899.1 GL888486 EGI60214.1 PNF37062.1 EFN71027.1 GFDF01005190 JAV08894.1 U69260 AAC34736.1 JXUM01044727 KQ561411 KXJ78663.1 AJWK01028468 EU124571 ABV60289.1 JXUM01028578 KQ560825 KXJ80731.1 U69261 AAC34737.1 EZ933291 ADJ57672.1 GAPW01004424 JAC09174.1 KYN21529.1 GU215629 ADD81447.1
EU137137 ABX39557.1 ODYU01006273 SOQ47964.1 EU137124 ABX39544.1 KZ150046 PZC74493.1 EU137125 ABX39545.1 KT984952 AMY63171.1 KQ460045 KPJ18126.1 EU137139 ABX39559.1 KQ459604 KPI92374.1 NWSH01003917 PCG65694.1 PCG65695.1 EU137138 ABX39558.1 EU265820 ABY88947.1 JTDY01000350 KOB77590.1 EU137119 ABX39539.1 EU137121 ABX39541.1 EU137122 ABX39542.1 EU137130 ABX39550.1 EU137128 ABX39548.1 EU137118 ABX39538.1 EU265822 ABY88949.1 AGBW02013167 OWR43916.1 EU137136 ABX39556.1 EU265819 ABY88946.1 EU137131 ABX39551.1 RSAL01000001 RVE55341.1 EU137117 ABX39537.1 EU137133 ABX39553.1 EU137135 ABX39555.1 EU137132 ABX39552.1 EU265818 ABY88945.1 EU137129 ABX39549.1 EU137126 ABX39546.1 EU265817 ABY88944.1 AY425622 AAR84202.1 EU137127 ABX39547.1 KK107077 EZA60591.1 QOIP01000001 EZA60607.1 RLU26326.1 RLU26700.1 EU137134 ABX39554.1 KK853323 KDR08524.1 RLU27030.1 GL768066 EFZ12622.1 GBYB01000663 JAG70430.1 PYGN01000874 PSN39804.1 GL437123 EFN71014.1 KQ414633 KOC67100.1 PYGN01000332 PSN48368.1 CH477978 CH477559 EAT34668.1 EAT38996.1 KQ982182 KYQ59195.1 EU031911 ABV44704.1 KQ981791 KYN35782.1 KQ977279 KYN04372.1 EAT38997.1 KZ288364 PBC26914.1 EF202179 ABP04045.1 JXUM01006083 KQ560208 KXJ83828.1 AF072220 AAD13531.1 GANO01001740 JAB58131.1 EAT34669.1 GFDF01005179 JAV08905.1 GFDF01005178 JAV08906.1 EZ933294 ADJ57675.1 KY661332 AVA17419.1 EAT38998.1 NEVH01006723 PNF37061.1 AF072222 AAD13533.1 U78970 AAC34312.1 GANO01000999 JAB58872.1 JXUM01006095 KXJ83830.1 ADTU01014245 KQ979440 KYN21516.1 U69957 AAB82404.1 GU301883 ADB92492.1 EAT34666.1 KQ435729 KOX77672.1 DS232267 EDS38794.1 KQ976464 KYM84538.1 EZA60593.1 GFDF01005185 JAV08899.1 GL888486 EGI60214.1 PNF37062.1 EFN71027.1 GFDF01005190 JAV08894.1 U69260 AAC34736.1 JXUM01044727 KQ561411 KXJ78663.1 AJWK01028468 EU124571 ABV60289.1 JXUM01028578 KQ560825 KXJ80731.1 U69261 AAC34737.1 EZ933291 ADJ57672.1 GAPW01004424 JAC09174.1 KYN21529.1 GU215629 ADD81447.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
+ More
UP000283053 UP000053097 UP000279307 UP000027135 UP000245037 UP000000311 UP000053825 UP000008820 UP000075809 UP000078541 UP000078542 UP000242457 UP000069940 UP000249989 UP000005203 UP000235965 UP000002358 UP000005205 UP000078492 UP000053105 UP000002320 UP000078540 UP000007755 UP000092461
UP000283053 UP000053097 UP000279307 UP000027135 UP000245037 UP000000311 UP000053825 UP000008820 UP000075809 UP000078541 UP000078542 UP000242457 UP000069940 UP000249989 UP000005203 UP000235965 UP000002358 UP000005205 UP000078492 UP000053105 UP000002320 UP000078540 UP000007755 UP000092461
PRIDE
Pfam
PF06757 Ins_allergen_rp
Interpro
IPR010629
Ins_allergen
ProteinModelPortal
H9J684
H9J683
B2KSG9
B2KSG6
B2KSI3
A0A2H1W637
+ More
B2KSH0 A0A2W1BRV7 B2KSH1 A0A161H9I2 A0A194RQ23 B2KSI5 A0A194PI21 A0A2A4J0T9 B2KSI4 B2KXE0 A0A0L7LQ68 B2KSG5 B2KSG7 B2KSG8 B2KSH6 B2KSH4 B2KSG4 B2KXE2 A0A212EQZ3 B2KSI2 B2KXD9 B2KSH7 A0A3S2LJN8 B2KSG3 B2KSH9 B2KSI1 B2KSH8 B2KXD8 B2KSH5 B2KSH2 B2KXD7 Q6TAW0 B2KSH3 A0A026WXX3 A0A026WX70 B2KSI0 A0A067QVM1 A0A3L8E2R5 E9J3M2 A0A0C9PIB4 A0A2P8Y6L1 E2A6C9 A0A0L7R895 A0A2P8YVU1 Q16K34 A0A151XFV9 A8C9U5 A0A151JU60 A0A195CV20 Q16WN9 A0A2A3E602 A9XFX0 A0A182G0S0 O96522 A0A088AIT4 U5EVT0 Q16K33 A0A1L8DRE9 A0A1L8DR00 F4MI46 A0A087ZXS1 A0A2P0XIZ6 Q16WN7 A0A2J7R8A2 Q9TZR6 O18535 U5EMJ5 A0A182G0S2 K7J3S2 A0A158NFC6 A0A195E8S7 O18530 D3JUE8 Q16K36 A0A0N0BIK3 B0X1R6 A0A195BI59 A0A026WXQ5 A0A1L8DR35 F4X027 A0A2J7R8A9 E2A6E2 A0A1L8DR95 O18527 A0A182G6A6 A8CW22 A0A182H7M5 A0A087ZXS3 O18528 F4MI43 A0A023ELN6 A0A195E9N4 D4NTK1
B2KSH0 A0A2W1BRV7 B2KSH1 A0A161H9I2 A0A194RQ23 B2KSI5 A0A194PI21 A0A2A4J0T9 B2KSI4 B2KXE0 A0A0L7LQ68 B2KSG5 B2KSG7 B2KSG8 B2KSH6 B2KSH4 B2KSG4 B2KXE2 A0A212EQZ3 B2KSI2 B2KXD9 B2KSH7 A0A3S2LJN8 B2KSG3 B2KSH9 B2KSI1 B2KSH8 B2KXD8 B2KSH5 B2KSH2 B2KXD7 Q6TAW0 B2KSH3 A0A026WXX3 A0A026WX70 B2KSI0 A0A067QVM1 A0A3L8E2R5 E9J3M2 A0A0C9PIB4 A0A2P8Y6L1 E2A6C9 A0A0L7R895 A0A2P8YVU1 Q16K34 A0A151XFV9 A8C9U5 A0A151JU60 A0A195CV20 Q16WN9 A0A2A3E602 A9XFX0 A0A182G0S0 O96522 A0A088AIT4 U5EVT0 Q16K33 A0A1L8DRE9 A0A1L8DR00 F4MI46 A0A087ZXS1 A0A2P0XIZ6 Q16WN7 A0A2J7R8A2 Q9TZR6 O18535 U5EMJ5 A0A182G0S2 K7J3S2 A0A158NFC6 A0A195E8S7 O18530 D3JUE8 Q16K36 A0A0N0BIK3 B0X1R6 A0A195BI59 A0A026WXQ5 A0A1L8DR35 F4X027 A0A2J7R8A9 E2A6E2 A0A1L8DR95 O18527 A0A182G6A6 A8CW22 A0A182H7M5 A0A087ZXS3 O18528 F4MI43 A0A023ELN6 A0A195E9N4 D4NTK1
PDB
4JRB
E-value=4.80414e-08,
Score=134
Ontologies
GO
PANTHER
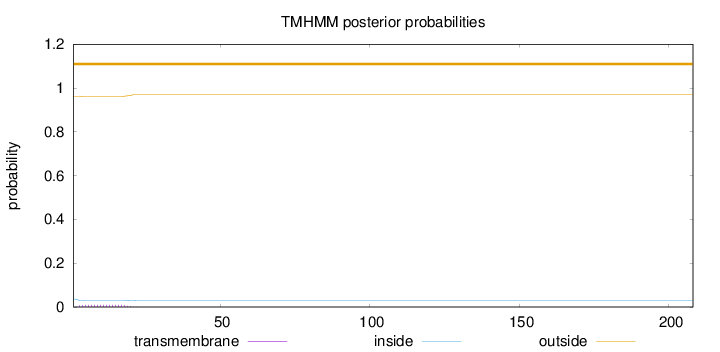
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.965287
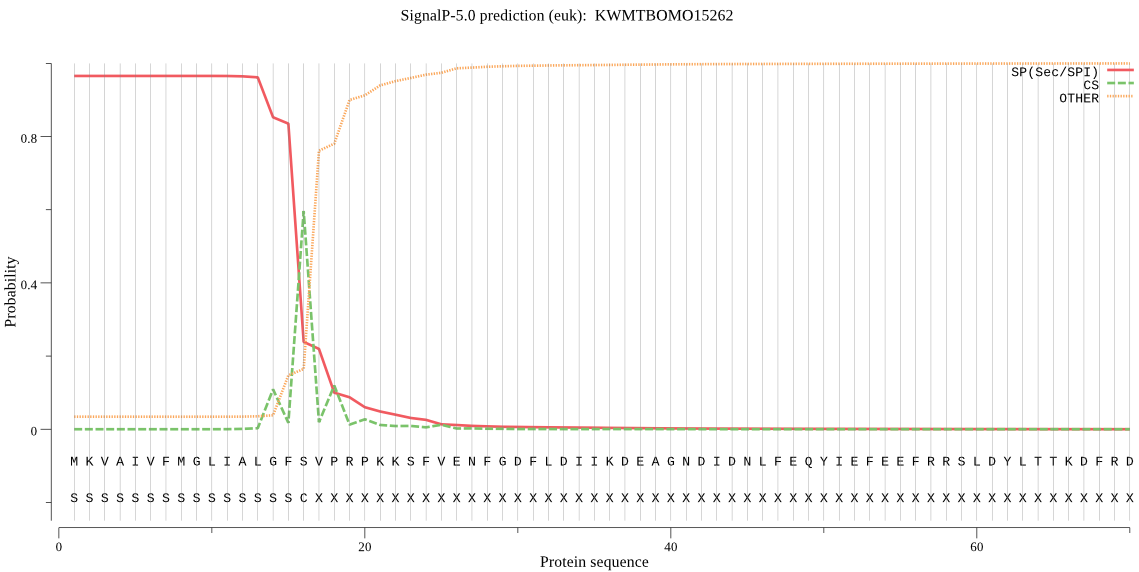
Length:
208
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16762
Exp number, first 60 AAs:
0.16743
Total prob of N-in:
0.03692
outside
1 - 208
Population Genetic Test Statistics
Pi
5.702163
Theta
13.210391
Tajima's D
-1.330279
CLR
0.033691
CSRT
0.08009599520024
Interpretation
Uncertain
