Gene
KWMTBOMO15259
Annotation
PREDICTED:_phosphatidylinositol_N-acetylglucosaminyltransferase_subunit_H_isoform_X2_[Bombyx_mori]
Full name
Phosphatidylinositol N-acetylglucosaminyltransferase subunit H
Alternative Name
Phosphatidylinositol-glycan biosynthesis class H protein
Location in the cell
Nuclear Reliability : 0.865
Sequence
CDS
ATGCTAAAATCAGAAACAGTCTTGATCGTTCCAAATGTTGGTATACGCAGCACTCAGAATTATCCAACCAGAAACTTACACACCTTTGTATCATGGGACAGAATTGAAGATATTATTATAAATGAAGTTATTGTGACGTCTAAAGTTCTTTATTACTTGACTATTTTGGTTAAAGAAGATCCTGAGAACACTGGAAACATTGAACAAAATCAATCAACCCGACTGGTGCCTCTTTTTTTGAGGACAAGACCCAGTTTGGCGGTGTTGGAGAAGATTTATGCAGATGTACAAGACTTACTGACGGAGGCTAAGCAACAAGTGTAA
Protein
MLKSETVLIVPNVGIRSTQNYPTRNLHTFVSWDRIEDIIINEVIVTSKVLYYLTILVKEDPENTGNIEQNQSTRLVPLFLRTRPSLAVLEKIYADVQDLLTEAKQQV
Summary
Description
Part of the complex catalyzing the transfer of N-acetylglucosamine from UDP-N-acetylglucosamine to phosphatidylinositol, the first step of GPI biosynthesis.
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol) + UDP-N-acetyl-alpha-D-glucosamine = a 6-(N-acetyl-alpha-D-glucosaminyl)-1-phosphatidyl-1D-myo-inositol + H(+) + UDP
Subunit
Associates with PIGA, PIGC, PIGP, PIGQ and DPM2. The latter is not essential for activity (By similarity).
Similarity
Belongs to the PIGH family.
Keywords
Complete proteome
Cytoplasm
Glycosyltransferase
Reference proteome
Transferase
Feature
chain Phosphatidylinositol N-acetylglucosaminyltransferase subunit H
Uniprot
A0A1E1VZA3
A0A194PGE1
A0A2H1WE54
A0A3S2NSN5
A0A2W1BLG0
A0A194RJX1
+ More
A0A2A4JA89 A0A212F711 A0A1A8M804 A0A1A8H187 A0A1A8CGW3 A0A1A8NBV4 A0A1A8U1K9 A0A1A8K412 A0A3Q1GBW4 A0A3B3QDI2 A0A3B4U5H9 A0A1A8S3R1 A0A1A7YIC8 H0XE05 A0A3B4W8V9 A0A3B3VBK0 A0A0S7LNY9 A0A3Q1B238 A0A3P8UA51 A0A3B5LQU0 A0A3B3Z0T3 M3ZX75 A0A087Y4H5 A0A3Q3M5P4 A0A2U9CT63 A0A096LUA4 A0A1A8IPH1 A0A315VWX9 I3IWG8 A0A3B4ZW30 A0A1U7QQY2 Q5M9N4 A0A0P4WBV5 A0A3Q2XN48 A0A2K5JTX7 W5N3H0 G3H1L9 A0A3B4F0N6 A0A3Q4I5K8 A0A3Q2W6A7 A0A3P8NYI5 A0A3P9C576 A0A3Q3K2V3 A0A3Q3B943 H0V0L7 A0A3B5K8G5 R0LCM1 A0A1A6G0X7 U3HZL7 D2HRG4 A0A212CTC5 A0A3Q3NDK9 A0A2Y9MYL5 A0A2Y9N9T7 A0A146ZN65 G5C7X7 S9WMG6 A0A2Y9N3L7 C1BXY9 A0A2U3V0Q8 A0A341BJW0 U6CTX9 M3Y6V1 L5M1N3 A0A2U4A9Y5 D4A0Z0 A0A3P9PE53 A0A3P9PE38 A0A2R8MDP4 A0A2Y9FFY8 A0A287A3K3 A0A2Y9DB08 A0A2R8PH55 A0A0E9X8D9 A0A3P9AH83 A0A3Q0QS54 A0A3P4LSH1 A0A2K5EK16 A0A091CZR3 U3CIG1 W5QJ53 A0A096NVP3 G8F4W5 F6URF9 A0A3P8YXB1 A0A2K5S1L1 F1MGP1 A0A2K6S5P0 A0A3L7HKN0 L8HYV5 A0A384AAP4 Q32L89 A0A0P5EBK7 A0A3Q7W581
A0A2A4JA89 A0A212F711 A0A1A8M804 A0A1A8H187 A0A1A8CGW3 A0A1A8NBV4 A0A1A8U1K9 A0A1A8K412 A0A3Q1GBW4 A0A3B3QDI2 A0A3B4U5H9 A0A1A8S3R1 A0A1A7YIC8 H0XE05 A0A3B4W8V9 A0A3B3VBK0 A0A0S7LNY9 A0A3Q1B238 A0A3P8UA51 A0A3B5LQU0 A0A3B3Z0T3 M3ZX75 A0A087Y4H5 A0A3Q3M5P4 A0A2U9CT63 A0A096LUA4 A0A1A8IPH1 A0A315VWX9 I3IWG8 A0A3B4ZW30 A0A1U7QQY2 Q5M9N4 A0A0P4WBV5 A0A3Q2XN48 A0A2K5JTX7 W5N3H0 G3H1L9 A0A3B4F0N6 A0A3Q4I5K8 A0A3Q2W6A7 A0A3P8NYI5 A0A3P9C576 A0A3Q3K2V3 A0A3Q3B943 H0V0L7 A0A3B5K8G5 R0LCM1 A0A1A6G0X7 U3HZL7 D2HRG4 A0A212CTC5 A0A3Q3NDK9 A0A2Y9MYL5 A0A2Y9N9T7 A0A146ZN65 G5C7X7 S9WMG6 A0A2Y9N3L7 C1BXY9 A0A2U3V0Q8 A0A341BJW0 U6CTX9 M3Y6V1 L5M1N3 A0A2U4A9Y5 D4A0Z0 A0A3P9PE53 A0A3P9PE38 A0A2R8MDP4 A0A2Y9FFY8 A0A287A3K3 A0A2Y9DB08 A0A2R8PH55 A0A0E9X8D9 A0A3P9AH83 A0A3Q0QS54 A0A3P4LSH1 A0A2K5EK16 A0A091CZR3 U3CIG1 W5QJ53 A0A096NVP3 G8F4W5 F6URF9 A0A3P8YXB1 A0A2K5S1L1 F1MGP1 A0A2K6S5P0 A0A3L7HKN0 L8HYV5 A0A384AAP4 Q32L89 A0A0P5EBK7 A0A3Q7W581
EC Number
2.4.1.198
Pubmed
EMBL
GDQN01010994
JAT80060.1
KQ459604
KPI92372.1
ODYU01008067
SOQ51349.1
+ More
RSAL01000001 RVE55343.1 KZ150046 PZC74495.1 KQ460045 KPJ18128.1 NWSH01002167 PCG69005.1 AGBW02009942 OWR49532.1 HAEF01011798 SBR52957.1 HAEB01009751 HAEC01009037 SBQ77253.1 HADZ01014367 HAEA01007840 SBP78308.1 HAEG01001976 SBR66334.1 HADY01011441 HAEJ01001503 SBS41960.1 HAEE01006937 SBR26957.1 HAEH01010132 HAEI01010994 SBS12174.1 HADW01018477 HADX01008105 SBP30337.1 AAQR03134929 GBYX01158480 JAO91174.1 AYCK01008713 CP026262 AWP19834.1 AWP19835.1 AWP19836.1 AWP19837.1 HAED01012781 SBQ99183.1 NHOQ01001000 PWA27820.1 AERX01005496 AK167892 BC086806 GDRN01075575 JAI63039.1 AHAT01019062 AHAT01019063 JH000107 EGV91866.1 AAKN02025210 AAKN02025211 KB743356 EOA99244.1 LZPO01107950 OBS59826.1 ADON01090394 ACTA01105694 ACTA01113694 GL193220 EFB27438.1 MKHE01000012 OWK09240.1 GCES01018533 JAR67790.1 JH173719 GEBF01003757 EHB17638.1 JAN99875.1 KB017142 EPY79727.1 BT079468 ACO13892.1 HAAF01002065 CCP73891.1 AEYP01004105 AEYP01004106 KB105175 ELK32554.1 AABR07064961 CH473947 EDM03717.1 AEMK02000061 DQIR01245654 DQIR01287508 HDC01132.1 HDC42986.1 GBXM01010457 JAH98120.1 CYRY02002172 VCW66832.1 KN123629 KFO24192.1 GAMT01007528 GAMS01004987 GAMR01002640 GAMQ01003428 JAB04333.1 JAB18149.1 JAB31292.1 JAB38423.1 AMGL01104303 AHZZ02032335 AQIA01065284 JH331132 EHH62329.1 JSUE03039944 JSUE03039945 CM001259 EHH27980.1 RAZU01000203 RLQ65891.1 JH882530 ELR49056.1 BC109701 GDIP01145304 JAJ78098.1
RSAL01000001 RVE55343.1 KZ150046 PZC74495.1 KQ460045 KPJ18128.1 NWSH01002167 PCG69005.1 AGBW02009942 OWR49532.1 HAEF01011798 SBR52957.1 HAEB01009751 HAEC01009037 SBQ77253.1 HADZ01014367 HAEA01007840 SBP78308.1 HAEG01001976 SBR66334.1 HADY01011441 HAEJ01001503 SBS41960.1 HAEE01006937 SBR26957.1 HAEH01010132 HAEI01010994 SBS12174.1 HADW01018477 HADX01008105 SBP30337.1 AAQR03134929 GBYX01158480 JAO91174.1 AYCK01008713 CP026262 AWP19834.1 AWP19835.1 AWP19836.1 AWP19837.1 HAED01012781 SBQ99183.1 NHOQ01001000 PWA27820.1 AERX01005496 AK167892 BC086806 GDRN01075575 JAI63039.1 AHAT01019062 AHAT01019063 JH000107 EGV91866.1 AAKN02025210 AAKN02025211 KB743356 EOA99244.1 LZPO01107950 OBS59826.1 ADON01090394 ACTA01105694 ACTA01113694 GL193220 EFB27438.1 MKHE01000012 OWK09240.1 GCES01018533 JAR67790.1 JH173719 GEBF01003757 EHB17638.1 JAN99875.1 KB017142 EPY79727.1 BT079468 ACO13892.1 HAAF01002065 CCP73891.1 AEYP01004105 AEYP01004106 KB105175 ELK32554.1 AABR07064961 CH473947 EDM03717.1 AEMK02000061 DQIR01245654 DQIR01287508 HDC01132.1 HDC42986.1 GBXM01010457 JAH98120.1 CYRY02002172 VCW66832.1 KN123629 KFO24192.1 GAMT01007528 GAMS01004987 GAMR01002640 GAMQ01003428 JAB04333.1 JAB18149.1 JAB31292.1 JAB38423.1 AMGL01104303 AHZZ02032335 AQIA01065284 JH331132 EHH62329.1 JSUE03039944 JSUE03039945 CM001259 EHH27980.1 RAZU01000203 RLQ65891.1 JH882530 ELR49056.1 BC109701 GDIP01145304 JAJ78098.1
Proteomes
UP000053268
UP000283053
UP000053240
UP000218220
UP000007151
UP000257200
+ More
UP000261540 UP000261420 UP000005225 UP000261360 UP000261500 UP000257160 UP000265080 UP000261380 UP000261480 UP000002852 UP000028760 UP000261640 UP000246464 UP000005207 UP000261400 UP000189706 UP000000589 UP000264820 UP000233080 UP000018468 UP000001075 UP000261460 UP000261580 UP000264840 UP000265100 UP000265160 UP000261600 UP000264800 UP000005447 UP000005226 UP000092124 UP000016666 UP000008912 UP000261660 UP000248483 UP000006813 UP000245320 UP000252040 UP000000715 UP000002494 UP000242638 UP000008225 UP000248484 UP000008227 UP000248480 UP000265140 UP000261340 UP000233020 UP000028990 UP000002356 UP000028761 UP000009130 UP000233100 UP000006718 UP000233040 UP000009136 UP000233220 UP000273346 UP000261681 UP000286642
UP000261540 UP000261420 UP000005225 UP000261360 UP000261500 UP000257160 UP000265080 UP000261380 UP000261480 UP000002852 UP000028760 UP000261640 UP000246464 UP000005207 UP000261400 UP000189706 UP000000589 UP000264820 UP000233080 UP000018468 UP000001075 UP000261460 UP000261580 UP000264840 UP000265100 UP000265160 UP000261600 UP000264800 UP000005447 UP000005226 UP000092124 UP000016666 UP000008912 UP000261660 UP000248483 UP000006813 UP000245320 UP000252040 UP000000715 UP000002494 UP000242638 UP000008225 UP000248484 UP000008227 UP000248480 UP000265140 UP000261340 UP000233020 UP000028990 UP000002356 UP000028761 UP000009130 UP000233100 UP000006718 UP000233040 UP000009136 UP000233220 UP000273346 UP000261681 UP000286642
Pfam
PF10181 PIG-H
Interpro
IPR019328
GPI-GlcNAc_Trfase_PIG-H_dom
ProteinModelPortal
A0A1E1VZA3
A0A194PGE1
A0A2H1WE54
A0A3S2NSN5
A0A2W1BLG0
A0A194RJX1
+ More
A0A2A4JA89 A0A212F711 A0A1A8M804 A0A1A8H187 A0A1A8CGW3 A0A1A8NBV4 A0A1A8U1K9 A0A1A8K412 A0A3Q1GBW4 A0A3B3QDI2 A0A3B4U5H9 A0A1A8S3R1 A0A1A7YIC8 H0XE05 A0A3B4W8V9 A0A3B3VBK0 A0A0S7LNY9 A0A3Q1B238 A0A3P8UA51 A0A3B5LQU0 A0A3B3Z0T3 M3ZX75 A0A087Y4H5 A0A3Q3M5P4 A0A2U9CT63 A0A096LUA4 A0A1A8IPH1 A0A315VWX9 I3IWG8 A0A3B4ZW30 A0A1U7QQY2 Q5M9N4 A0A0P4WBV5 A0A3Q2XN48 A0A2K5JTX7 W5N3H0 G3H1L9 A0A3B4F0N6 A0A3Q4I5K8 A0A3Q2W6A7 A0A3P8NYI5 A0A3P9C576 A0A3Q3K2V3 A0A3Q3B943 H0V0L7 A0A3B5K8G5 R0LCM1 A0A1A6G0X7 U3HZL7 D2HRG4 A0A212CTC5 A0A3Q3NDK9 A0A2Y9MYL5 A0A2Y9N9T7 A0A146ZN65 G5C7X7 S9WMG6 A0A2Y9N3L7 C1BXY9 A0A2U3V0Q8 A0A341BJW0 U6CTX9 M3Y6V1 L5M1N3 A0A2U4A9Y5 D4A0Z0 A0A3P9PE53 A0A3P9PE38 A0A2R8MDP4 A0A2Y9FFY8 A0A287A3K3 A0A2Y9DB08 A0A2R8PH55 A0A0E9X8D9 A0A3P9AH83 A0A3Q0QS54 A0A3P4LSH1 A0A2K5EK16 A0A091CZR3 U3CIG1 W5QJ53 A0A096NVP3 G8F4W5 F6URF9 A0A3P8YXB1 A0A2K5S1L1 F1MGP1 A0A2K6S5P0 A0A3L7HKN0 L8HYV5 A0A384AAP4 Q32L89 A0A0P5EBK7 A0A3Q7W581
A0A2A4JA89 A0A212F711 A0A1A8M804 A0A1A8H187 A0A1A8CGW3 A0A1A8NBV4 A0A1A8U1K9 A0A1A8K412 A0A3Q1GBW4 A0A3B3QDI2 A0A3B4U5H9 A0A1A8S3R1 A0A1A7YIC8 H0XE05 A0A3B4W8V9 A0A3B3VBK0 A0A0S7LNY9 A0A3Q1B238 A0A3P8UA51 A0A3B5LQU0 A0A3B3Z0T3 M3ZX75 A0A087Y4H5 A0A3Q3M5P4 A0A2U9CT63 A0A096LUA4 A0A1A8IPH1 A0A315VWX9 I3IWG8 A0A3B4ZW30 A0A1U7QQY2 Q5M9N4 A0A0P4WBV5 A0A3Q2XN48 A0A2K5JTX7 W5N3H0 G3H1L9 A0A3B4F0N6 A0A3Q4I5K8 A0A3Q2W6A7 A0A3P8NYI5 A0A3P9C576 A0A3Q3K2V3 A0A3Q3B943 H0V0L7 A0A3B5K8G5 R0LCM1 A0A1A6G0X7 U3HZL7 D2HRG4 A0A212CTC5 A0A3Q3NDK9 A0A2Y9MYL5 A0A2Y9N9T7 A0A146ZN65 G5C7X7 S9WMG6 A0A2Y9N3L7 C1BXY9 A0A2U3V0Q8 A0A341BJW0 U6CTX9 M3Y6V1 L5M1N3 A0A2U4A9Y5 D4A0Z0 A0A3P9PE53 A0A3P9PE38 A0A2R8MDP4 A0A2Y9FFY8 A0A287A3K3 A0A2Y9DB08 A0A2R8PH55 A0A0E9X8D9 A0A3P9AH83 A0A3Q0QS54 A0A3P4LSH1 A0A2K5EK16 A0A091CZR3 U3CIG1 W5QJ53 A0A096NVP3 G8F4W5 F6URF9 A0A3P8YXB1 A0A2K5S1L1 F1MGP1 A0A2K6S5P0 A0A3L7HKN0 L8HYV5 A0A384AAP4 Q32L89 A0A0P5EBK7 A0A3Q7W581
Ontologies
PATHWAY
GO
Topology
Subcellular location
Cytoplasm
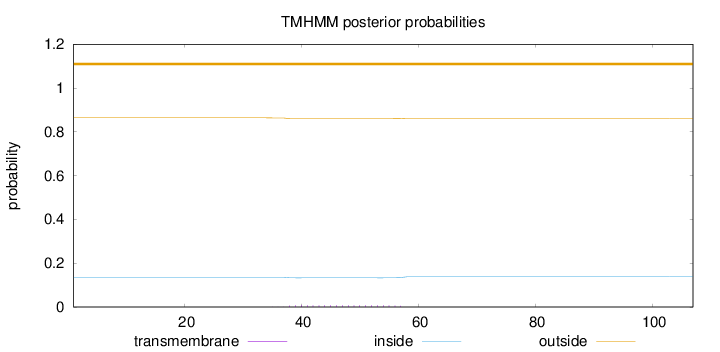
Length:
107
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14208
Exp number, first 60 AAs:
0.14208
Total prob of N-in:
0.13588
outside
1 - 107
Population Genetic Test Statistics
Pi
251.701613
Theta
144.644394
Tajima's D
2.322303
CLR
0.101175
CSRT
0.926803659817009
Interpretation
Uncertain
