Gene
KWMTBOMO15252
Annotation
PREDICTED:_nose_resistant_to_fluoxetine_protein_6-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.897
Sequence
CDS
ATGCAAAGGCAAAGGGGGGAGAGTGAAGCGCTCGTAGCAGCACTTTCAGTGGTAGTGTTCTATGTTTCTGCTGTATACCCTTACACCGGCACCGGCCCATTGTGGAACAAAGCAATCGCTGCTGAAACAGAAGCATGCCGTAAGAACTGGTGGCTTAATCTACTCATGATCAGCAATTATGTTGACACCGAAAATATTTGCATCGTAATATCCTGGTACATTCCGTGCGACTTCCACTTTTTCTTGGTCACAATTATTGGATTTTTTTTATACAAGAAGTACCCGAAAATAGGCGTTATCACCGGAGGAGTATTGTTTGTGGTCGCACTATTGATACCGGGCATCATGACGTACATGTACCAGTTGGCTGCAGTACAACTGTTTACTATTGAGTGA
Protein
MQRQRGESEALVAALSVVVFYVSAVYPYTGTGPLWNKAIAAETEACRKNWWLNLLMISNYVDTENICIVISWYIPCDFHFFLVTIIGFFLYKKYPKIGVITGGVLFVVALLIPGIMTYMYQLAAVQLFTIE
Summary
Uniprot
H9J678
A0A2A4IZN6
A0A2W1BP43
A0A0L7LIX9
A0A194RL37
A0A2H1VF27
+ More
A0A3S2LZ61 A0A212F1J8 A0A194QEM0 A0A2A4JJD7 U4UQL1 N6U8Y8 A0A1B6JH26 A0A1E1WGX5 A0A2H1W229 A0A1E1WQD5 D6WQE4 A0A1E1WB37 A0A1E1WUR1 A0A0N0PE46 A0A1E1W9B1 A0A1E1WIK4 A0A1B6KDE2 N6UHN2 A0A1B6LPM9 A0A2H8U115 A0A1B6HSS5 A0A2S2PXA7 A0A2J7QPC5 A0A2J7QP78 A0A2A4JJ41 N6TZ73 U4U7K4 A0A2S2QG12 A0A1B6CQA8 A0A0N1PH40 A0A1Z5L7Q4 A0A1S4EQR7 J9LI86 A0A2S2Q1M5 A0A1Y1M2Q0 A0A2J7Q6G4 A0A3Q0JA99 A0A2S2Q8L1 A0A2H1W4H1 A0A2S2NPC3 A0A1W4X317 J9JKB2 A0A139W8X9 H9JNJ3 A0A182YEG1 A0A2A4JXW1 A0A2H8U041 A0A3S2LST9 A0A2S2P4J7 W5JUV2 A0A182WLD5 A0A2H1W868 J9LI88 A0A2P8XRP1 A0A026WGT0 A0A2S2QA26 A0A3Q0ISA9 A0A2S2QQV9 A0A131YTH9 A0A2S2QKZ1 A0A232FMR4 A0A2S2NYS9 A0A084W2G5 A0A182W4K6 A0A2M4CX01 A0A2J7QG24 A0A182I2D7 J9KT87 A0A182VP89 A0A182KQ82 A0A087SXH8 A0A182JDV2 A0A182MV59 A0A182UKV1 A0A1B6KSX3 A0A182TKI8 A0A182K0Z6
A0A3S2LZ61 A0A212F1J8 A0A194QEM0 A0A2A4JJD7 U4UQL1 N6U8Y8 A0A1B6JH26 A0A1E1WGX5 A0A2H1W229 A0A1E1WQD5 D6WQE4 A0A1E1WB37 A0A1E1WUR1 A0A0N0PE46 A0A1E1W9B1 A0A1E1WIK4 A0A1B6KDE2 N6UHN2 A0A1B6LPM9 A0A2H8U115 A0A1B6HSS5 A0A2S2PXA7 A0A2J7QPC5 A0A2J7QP78 A0A2A4JJ41 N6TZ73 U4U7K4 A0A2S2QG12 A0A1B6CQA8 A0A0N1PH40 A0A1Z5L7Q4 A0A1S4EQR7 J9LI86 A0A2S2Q1M5 A0A1Y1M2Q0 A0A2J7Q6G4 A0A3Q0JA99 A0A2S2Q8L1 A0A2H1W4H1 A0A2S2NPC3 A0A1W4X317 J9JKB2 A0A139W8X9 H9JNJ3 A0A182YEG1 A0A2A4JXW1 A0A2H8U041 A0A3S2LST9 A0A2S2P4J7 W5JUV2 A0A182WLD5 A0A2H1W868 J9LI88 A0A2P8XRP1 A0A026WGT0 A0A2S2QA26 A0A3Q0ISA9 A0A2S2QQV9 A0A131YTH9 A0A2S2QKZ1 A0A232FMR4 A0A2S2NYS9 A0A084W2G5 A0A182W4K6 A0A2M4CX01 A0A2J7QG24 A0A182I2D7 J9KT87 A0A182VP89 A0A182KQ82 A0A087SXH8 A0A182JDV2 A0A182MV59 A0A182UKV1 A0A1B6KSX3 A0A182TKI8 A0A182K0Z6
Pubmed
EMBL
BABH01019947
BABH01019948
BABH01019949
BABH01019950
NWSH01004589
PCG64888.1
+ More
KZ150046 PZC74500.1 JTDY01000928 KOB75402.1 KQ460045 KPJ18134.1 ODYU01002228 SOQ39435.1 RSAL01000102 RVE47464.1 AGBW02010872 OWR47594.1 KQ459054 KPJ03927.1 NWSH01001289 PCG71838.1 KB632326 ERL92431.1 APGK01035013 APGK01035014 APGK01035015 KB740923 ENN78145.1 GECU01009231 JAS98475.1 GDQN01011399 GDQN01004801 JAT79655.1 JAT86253.1 ODYU01005850 SOQ47141.1 GDQN01001866 JAT89188.1 KQ971354 EFA06959.1 GDQN01006834 JAT84220.1 GDQN01000299 JAT90755.1 KQ460047 KPJ17986.1 GDQN01007466 JAT83588.1 GDQN01004215 JAT86839.1 GEBQ01030520 JAT09457.1 APGK01035017 APGK01035018 APGK01035019 APGK01035020 ENN78147.1 GEBQ01014418 JAT25559.1 GFXV01007717 MBW19522.1 GECU01029990 GECU01001596 JAS77716.1 JAT06111.1 GGMS01000820 MBY70023.1 NEVH01012096 PNF30393.1 PNF30392.1 PCG71839.1 APGK01049501 APGK01049502 KB741155 ENN73651.1 KB630423 KB631761 ERL83595.1 ERL85915.1 GGMS01007461 MBY76664.1 GEDC01021708 JAS15590.1 KPJ17987.1 GFJQ02003514 JAW03456.1 ABLF02032424 ABLF02032427 GGMS01002340 MBY71543.1 GEZM01042633 JAV79751.1 NEVH01017478 PNF24171.1 GGMS01004866 MBY74069.1 ODYU01006284 SOQ47989.1 GGMR01006421 MBY19040.1 ABLF02037491 ABLF02037494 ABLF02037498 ABLF02037500 ABLF02037506 KQ972879 KXZ75738.1 BABH01041997 NWSH01000450 PCG76333.1 GFXV01007317 MBW19122.1 RSAL01000275 RVE43113.1 GGMR01011802 MBY24421.1 ADMH02000427 ETN66544.1 ODYU01006653 SOQ48684.1 ABLF02004907 ABLF02004908 ABLF02004909 ABLF02004910 ABLF02031628 ABLF02031630 PYGN01001466 PSN34669.1 KK107213 EZA55272.1 GGMS01005383 MBY74586.1 GGMS01010891 MBY80094.1 GEDV01006712 JAP81845.1 GGMS01009213 MBY78416.1 NNAY01000028 OXU31810.1 GGMR01009762 MBY22381.1 ATLV01019615 KE525275 KFB44409.1 GGFL01005669 MBW69847.1 NEVH01014836 PNF27544.1 APCN01000146 ABLF02022069 KK112404 KFM57567.1 AXCM01001275 GEBQ01025424 JAT14553.1
KZ150046 PZC74500.1 JTDY01000928 KOB75402.1 KQ460045 KPJ18134.1 ODYU01002228 SOQ39435.1 RSAL01000102 RVE47464.1 AGBW02010872 OWR47594.1 KQ459054 KPJ03927.1 NWSH01001289 PCG71838.1 KB632326 ERL92431.1 APGK01035013 APGK01035014 APGK01035015 KB740923 ENN78145.1 GECU01009231 JAS98475.1 GDQN01011399 GDQN01004801 JAT79655.1 JAT86253.1 ODYU01005850 SOQ47141.1 GDQN01001866 JAT89188.1 KQ971354 EFA06959.1 GDQN01006834 JAT84220.1 GDQN01000299 JAT90755.1 KQ460047 KPJ17986.1 GDQN01007466 JAT83588.1 GDQN01004215 JAT86839.1 GEBQ01030520 JAT09457.1 APGK01035017 APGK01035018 APGK01035019 APGK01035020 ENN78147.1 GEBQ01014418 JAT25559.1 GFXV01007717 MBW19522.1 GECU01029990 GECU01001596 JAS77716.1 JAT06111.1 GGMS01000820 MBY70023.1 NEVH01012096 PNF30393.1 PNF30392.1 PCG71839.1 APGK01049501 APGK01049502 KB741155 ENN73651.1 KB630423 KB631761 ERL83595.1 ERL85915.1 GGMS01007461 MBY76664.1 GEDC01021708 JAS15590.1 KPJ17987.1 GFJQ02003514 JAW03456.1 ABLF02032424 ABLF02032427 GGMS01002340 MBY71543.1 GEZM01042633 JAV79751.1 NEVH01017478 PNF24171.1 GGMS01004866 MBY74069.1 ODYU01006284 SOQ47989.1 GGMR01006421 MBY19040.1 ABLF02037491 ABLF02037494 ABLF02037498 ABLF02037500 ABLF02037506 KQ972879 KXZ75738.1 BABH01041997 NWSH01000450 PCG76333.1 GFXV01007317 MBW19122.1 RSAL01000275 RVE43113.1 GGMR01011802 MBY24421.1 ADMH02000427 ETN66544.1 ODYU01006653 SOQ48684.1 ABLF02004907 ABLF02004908 ABLF02004909 ABLF02004910 ABLF02031628 ABLF02031630 PYGN01001466 PSN34669.1 KK107213 EZA55272.1 GGMS01005383 MBY74586.1 GGMS01010891 MBY80094.1 GEDV01006712 JAP81845.1 GGMS01009213 MBY78416.1 NNAY01000028 OXU31810.1 GGMR01009762 MBY22381.1 ATLV01019615 KE525275 KFB44409.1 GGFL01005669 MBW69847.1 NEVH01014836 PNF27544.1 APCN01000146 ABLF02022069 KK112404 KFM57567.1 AXCM01001275 GEBQ01025424 JAT14553.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000283053
UP000007151
+ More
UP000053268 UP000030742 UP000019118 UP000007266 UP000235965 UP000079169 UP000007819 UP000192223 UP000076408 UP000000673 UP000075920 UP000245037 UP000053097 UP000215335 UP000030765 UP000075840 UP000075903 UP000075882 UP000054359 UP000075880 UP000075883 UP000075902 UP000075881
UP000053268 UP000030742 UP000019118 UP000007266 UP000235965 UP000079169 UP000007819 UP000192223 UP000076408 UP000000673 UP000075920 UP000245037 UP000053097 UP000215335 UP000030765 UP000075840 UP000075903 UP000075882 UP000054359 UP000075880 UP000075883 UP000075902 UP000075881
Interpro
ProteinModelPortal
H9J678
A0A2A4IZN6
A0A2W1BP43
A0A0L7LIX9
A0A194RL37
A0A2H1VF27
+ More
A0A3S2LZ61 A0A212F1J8 A0A194QEM0 A0A2A4JJD7 U4UQL1 N6U8Y8 A0A1B6JH26 A0A1E1WGX5 A0A2H1W229 A0A1E1WQD5 D6WQE4 A0A1E1WB37 A0A1E1WUR1 A0A0N0PE46 A0A1E1W9B1 A0A1E1WIK4 A0A1B6KDE2 N6UHN2 A0A1B6LPM9 A0A2H8U115 A0A1B6HSS5 A0A2S2PXA7 A0A2J7QPC5 A0A2J7QP78 A0A2A4JJ41 N6TZ73 U4U7K4 A0A2S2QG12 A0A1B6CQA8 A0A0N1PH40 A0A1Z5L7Q4 A0A1S4EQR7 J9LI86 A0A2S2Q1M5 A0A1Y1M2Q0 A0A2J7Q6G4 A0A3Q0JA99 A0A2S2Q8L1 A0A2H1W4H1 A0A2S2NPC3 A0A1W4X317 J9JKB2 A0A139W8X9 H9JNJ3 A0A182YEG1 A0A2A4JXW1 A0A2H8U041 A0A3S2LST9 A0A2S2P4J7 W5JUV2 A0A182WLD5 A0A2H1W868 J9LI88 A0A2P8XRP1 A0A026WGT0 A0A2S2QA26 A0A3Q0ISA9 A0A2S2QQV9 A0A131YTH9 A0A2S2QKZ1 A0A232FMR4 A0A2S2NYS9 A0A084W2G5 A0A182W4K6 A0A2M4CX01 A0A2J7QG24 A0A182I2D7 J9KT87 A0A182VP89 A0A182KQ82 A0A087SXH8 A0A182JDV2 A0A182MV59 A0A182UKV1 A0A1B6KSX3 A0A182TKI8 A0A182K0Z6
A0A3S2LZ61 A0A212F1J8 A0A194QEM0 A0A2A4JJD7 U4UQL1 N6U8Y8 A0A1B6JH26 A0A1E1WGX5 A0A2H1W229 A0A1E1WQD5 D6WQE4 A0A1E1WB37 A0A1E1WUR1 A0A0N0PE46 A0A1E1W9B1 A0A1E1WIK4 A0A1B6KDE2 N6UHN2 A0A1B6LPM9 A0A2H8U115 A0A1B6HSS5 A0A2S2PXA7 A0A2J7QPC5 A0A2J7QP78 A0A2A4JJ41 N6TZ73 U4U7K4 A0A2S2QG12 A0A1B6CQA8 A0A0N1PH40 A0A1Z5L7Q4 A0A1S4EQR7 J9LI86 A0A2S2Q1M5 A0A1Y1M2Q0 A0A2J7Q6G4 A0A3Q0JA99 A0A2S2Q8L1 A0A2H1W4H1 A0A2S2NPC3 A0A1W4X317 J9JKB2 A0A139W8X9 H9JNJ3 A0A182YEG1 A0A2A4JXW1 A0A2H8U041 A0A3S2LST9 A0A2S2P4J7 W5JUV2 A0A182WLD5 A0A2H1W868 J9LI88 A0A2P8XRP1 A0A026WGT0 A0A2S2QA26 A0A3Q0ISA9 A0A2S2QQV9 A0A131YTH9 A0A2S2QKZ1 A0A232FMR4 A0A2S2NYS9 A0A084W2G5 A0A182W4K6 A0A2M4CX01 A0A2J7QG24 A0A182I2D7 J9KT87 A0A182VP89 A0A182KQ82 A0A087SXH8 A0A182JDV2 A0A182MV59 A0A182UKV1 A0A1B6KSX3 A0A182TKI8 A0A182K0Z6
Ontologies
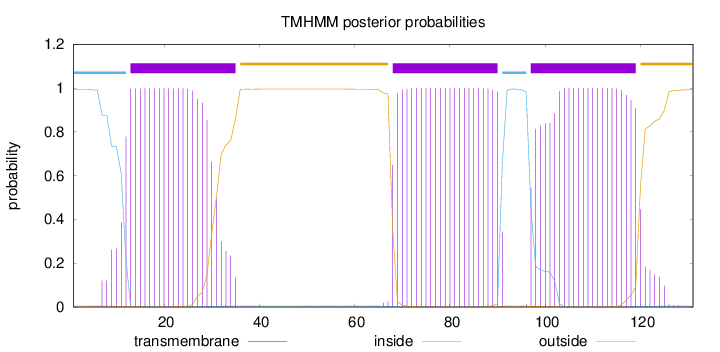
Topology
Length:
131
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
66.47869
Exp number, first 60 AAs:
20.73412
Total prob of N-in:
0.99505
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 67
TMhelix
68 - 90
inside
91 - 96
TMhelix
97 - 119
outside
120 - 131
Population Genetic Test Statistics
Pi
152.825807
Theta
157.905593
Tajima's D
-0.333816
CLR
46.564886
CSRT
0.286335683215839
Interpretation
Uncertain
