Gene
KWMTBOMO15250
Pre Gene Modal
BGIBMGA005017
Annotation
PREDICTED:_nose_resistant_to_fluoxetine_protein_6-like_[Bombyx_mori]
Full name
Nose resistant to fluoxetine protein 6
Location in the cell
PlasmaMembrane Reliability : 3.811
Sequence
CDS
ATGTTAAGTACACACACATATTATCACCCTCCCCACACGCGTCCTCTTAACGAGTTAACATGGGGAGTTTGCACTCCAGTTTCCTGTCTGCCCAGCACGATTGAGAAATTGATGGCCGTGACGTTGGCAAGGAGTCACCTCGGTGCAGCCGGTCTGAAAGCGAGGATTGCAGTTAATGAAAAATGTCAAACCAATGAAGATCAGAGTTACGATGGATTGTTTTATTCTTTTGTAATTTTAATGGGCTCCTTAATTTCGATTTGCTTAGTGTGCACTTATTACAACTGTACAAAACAAATTGAATCTAATTCAGCAAGTTCACATTTAATCAGAGCGTTTTGCATGAAAGAAAACGCTTCTGACCTATTGAAAATGAGAAAAGACGGATCAGAAGTAATTTACGGAATCAAATGGCTAACGATGTGTTTGATAGTTGTGGATCATCAAATCGGTATTGGTAATGCGGGTCCCATCAGCAACGGACTGACCTCAGATAATATAATTCAGTCTTTTTTGGGACTATTTATTCTACACGACGACTTATTTGTGGATACGTTTTTTCTTTTATCTGGCTTCCTAACGATTTCGACTTTCGCATCACTGAAAAAGCTGCCGAACCCGTTTATAGTGATCTTGAGAAGATACATCAGAAGGAAGACACCATACATACAGACGCCAATACCAACATCCGTCAGAAGCGGCAAAAGGCTTGACAAGTCTATAAGCATGTGA
Protein
MLSTHTYYHPPHTRPLNELTWGVCTPVSCLPSTIEKLMAVTLARSHLGAAGLKARIAVNEKCQTNEDQSYDGLFYSFVILMGSLISICLVCTYYNCTKQIESNSASSHLIRAFCMKENASDLLKMRKDGSEVIYGIKWLTMCLIVVDHQIGIGNAGPISNGLTSDNIIQSFLGLFILHDDLFVDTFFLLSGFLTISTFASLKKLPNPFIVILRRYIRRKTPYIQTPIPTSVRSGKRLDKSISM
Summary
Description
Plays a role in the uptake of a range of molecules including lipids and xenobiotic compounds from the intestine to surrounding tissues. Mediates transport of lipids from intestine to the reproductive tract. Required for efficient yolk transport into oocytes. Vital for embryonic development.
Similarity
Belongs to the acyltransferase 3 family.
Keywords
Complete proteome
Developmental protein
Glycoprotein
Lipid transport
Lipid-binding
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Transport
Feature
chain Nose resistant to fluoxetine protein 6
Uniprot
A0A2A4IZN6
A0A2W1BP43
A0A0L7LIX9
A0A2H1VF27
A0A194RL37
A0A194PG52
+ More
A0A3S2LZ61 A0A212F1J8 H9J678 A0A2A4JXW1 A0A2H1VCF8 A0A1E1WQD5 A0A1E1WB37 A0A1E1W9B1 A0A1E1WIK4 A0A2H1VRN7 A0A2H1WD69 A0A0N0PE46 N6U8Y8 A0A194QEM0 A0A2H1W229 A0A2H1V1A7 D6WQE4 B3MQQ0 A0A1E1WGX5 A0A1E1WUR1 A0A2H1UZX8 A0A2A4JJD7 A0A1B6CQA8 A0A2A4JY64 A0A194PEK6 K7J4D4 A0A2P8YMN8 A0A232EIQ2 Q09225 A0A1W4VFZ1 A0A194R3W4 J9KT87 A0A2H1W4H1 A0A1I7UQZ1 B4NAC3 B5DL87 A0A182U2H9 B4PZG5 B4I773 A0A2J7PZI3 B4M0Z0 A0A0M4FAS0 A0A1J1HH25 A0A182JM14 A0A3B0IZN8 Q9VWG5 A0A195BMB6 A0A1B0D242 A0A0V0GAI0 B4R216 A0A146LAZ8 A0A2A4JJ41 B4R7R7 B4HAQ3 A0A2H2IGP4 A0A182V747 A0A182IAC3 A0A182XEF8 A0A2P4WDN8 E3LGZ1 A0A224XDS6 A0A1B6J389 A0A232ELQ7 B4PZG6 A0A158NXG1 A0A151IFH6 E2A1E7 A0A195ECF1 A0NCC8 B4JME3 V5FVL9 A0A2P8Y4A3 A0A1B6M8Z4 A0A2A2J3J9 A0A261B190 A0A182K8B2 A8XHZ3 A0A1J1I9K6 D1ZZM5 U4UJ36 F4WQ68 A0A2A4J2E4 A0A2G5UXB0 W8BM55 A0A2H1WFD8 K7J4D6 A0A2J7PZF1 B4PNU7 A0A2S2QA26
A0A3S2LZ61 A0A212F1J8 H9J678 A0A2A4JXW1 A0A2H1VCF8 A0A1E1WQD5 A0A1E1WB37 A0A1E1W9B1 A0A1E1WIK4 A0A2H1VRN7 A0A2H1WD69 A0A0N0PE46 N6U8Y8 A0A194QEM0 A0A2H1W229 A0A2H1V1A7 D6WQE4 B3MQQ0 A0A1E1WGX5 A0A1E1WUR1 A0A2H1UZX8 A0A2A4JJD7 A0A1B6CQA8 A0A2A4JY64 A0A194PEK6 K7J4D4 A0A2P8YMN8 A0A232EIQ2 Q09225 A0A1W4VFZ1 A0A194R3W4 J9KT87 A0A2H1W4H1 A0A1I7UQZ1 B4NAC3 B5DL87 A0A182U2H9 B4PZG5 B4I773 A0A2J7PZI3 B4M0Z0 A0A0M4FAS0 A0A1J1HH25 A0A182JM14 A0A3B0IZN8 Q9VWG5 A0A195BMB6 A0A1B0D242 A0A0V0GAI0 B4R216 A0A146LAZ8 A0A2A4JJ41 B4R7R7 B4HAQ3 A0A2H2IGP4 A0A182V747 A0A182IAC3 A0A182XEF8 A0A2P4WDN8 E3LGZ1 A0A224XDS6 A0A1B6J389 A0A232ELQ7 B4PZG6 A0A158NXG1 A0A151IFH6 E2A1E7 A0A195ECF1 A0NCC8 B4JME3 V5FVL9 A0A2P8Y4A3 A0A1B6M8Z4 A0A2A2J3J9 A0A261B190 A0A182K8B2 A8XHZ3 A0A1J1I9K6 D1ZZM5 U4UJ36 F4WQ68 A0A2A4J2E4 A0A2G5UXB0 W8BM55 A0A2H1WFD8 K7J4D6 A0A2J7PZF1 B4PNU7 A0A2S2QA26
Pubmed
28756777
26227816
26354079
22118469
19121390
23537049
+ More
18362917 19820115 17994087 20075255 29403074 28648823 10488330 9851916 16487504 16118202 17761667 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26823975 26114425 26394399 21347285 20798317 12364791 14747013 17210077 14624247 21719571 24495485
18362917 19820115 17994087 20075255 29403074 28648823 10488330 9851916 16487504 16118202 17761667 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26823975 26114425 26394399 21347285 20798317 12364791 14747013 17210077 14624247 21719571 24495485
EMBL
NWSH01004589
PCG64888.1
KZ150046
PZC74500.1
JTDY01000928
KOB75402.1
+ More
ODYU01002228 SOQ39435.1 KQ460045 KPJ18134.1 KQ459604 KPI92366.1 RSAL01000102 RVE47464.1 AGBW02010872 OWR47594.1 BABH01019947 BABH01019948 BABH01019949 BABH01019950 NWSH01000450 PCG76333.1 ODYU01001799 SOQ38495.1 GDQN01001866 JAT89188.1 GDQN01006834 JAT84220.1 GDQN01007466 JAT83588.1 GDQN01004215 JAT86839.1 ODYU01004040 SOQ43513.1 ODYU01007817 SOQ50906.1 KQ460047 KPJ17986.1 APGK01035013 APGK01035014 APGK01035015 KB740923 ENN78145.1 KQ459054 KPJ03927.1 ODYU01005850 SOQ47141.1 ODYU01000038 SOQ34132.1 KQ971354 EFA06959.1 CH902621 EDV44676.1 GDQN01011399 GDQN01004801 JAT79655.1 JAT86253.1 GDQN01000299 JAT90755.1 ODYU01000040 SOQ34141.1 NWSH01001289 PCG71838.1 GEDC01021708 JAS15590.1 PCG76332.1 KQ459606 KPI91124.1 PYGN01000490 PSN45474.1 NNAY01004216 OXU18191.1 AF173372 Z46676 KQ460947 KPJ10531.1 ABLF02022069 ODYU01006284 SOQ47989.1 CH964232 EDW80737.1 CH379063 EDY72286.1 CM000162 EDX02121.1 CH480823 EDW56171.1 NEVH01020335 PNF21736.1 CH940650 EDW67401.2 CP012528 ALC49557.1 CVRI01000004 CRK87301.1 OUUW01000001 SPP73715.1 AE014298 AAF48976.1 KQ976441 KYM86395.1 AJVK01002698 AJVK01002699 AJVK01002700 GECL01000986 JAP05138.1 CM000364 EDX14073.1 GDHC01013884 JAQ04745.1 PCG71839.1 CM000366 EDX18400.1 CH479242 EDW37664.1 APCN01001143 LFJK02000006 POM47667.1 DS268408 EFO86322.1 GFTR01007282 JAW09144.1 GECU01014041 JAS93665.1 NNAY01003533 OXU19242.1 EDX02122.2 ADTU01003043 KQ977780 KYM99948.1 GL435766 EFN72620.1 KQ979074 KYN22900.1 AAAB01008846 EAU77249.3 CH916371 EDV92468.1 GALX01006776 JAB61690.1 PYGN01000947 PSN39086.1 GEBQ01007577 JAT32400.1 LIAE01010705 PAV56380.1 NMWX01000004 OZG03476.1 HE600919 CAP32259.2 CVRI01000044 CRK96891.1 KQ971338 EFA01831.2 KB632326 ERL92433.1 GL888262 EGI63691.1 NWSH01003464 PCG66235.1 PDUG01000002 PIC44143.1 GAMC01012119 JAB94436.1 ODYU01008317 SOQ51800.1 PNF21711.1 CM000160 EDW98157.2 GGMS01005383 MBY74586.1
ODYU01002228 SOQ39435.1 KQ460045 KPJ18134.1 KQ459604 KPI92366.1 RSAL01000102 RVE47464.1 AGBW02010872 OWR47594.1 BABH01019947 BABH01019948 BABH01019949 BABH01019950 NWSH01000450 PCG76333.1 ODYU01001799 SOQ38495.1 GDQN01001866 JAT89188.1 GDQN01006834 JAT84220.1 GDQN01007466 JAT83588.1 GDQN01004215 JAT86839.1 ODYU01004040 SOQ43513.1 ODYU01007817 SOQ50906.1 KQ460047 KPJ17986.1 APGK01035013 APGK01035014 APGK01035015 KB740923 ENN78145.1 KQ459054 KPJ03927.1 ODYU01005850 SOQ47141.1 ODYU01000038 SOQ34132.1 KQ971354 EFA06959.1 CH902621 EDV44676.1 GDQN01011399 GDQN01004801 JAT79655.1 JAT86253.1 GDQN01000299 JAT90755.1 ODYU01000040 SOQ34141.1 NWSH01001289 PCG71838.1 GEDC01021708 JAS15590.1 PCG76332.1 KQ459606 KPI91124.1 PYGN01000490 PSN45474.1 NNAY01004216 OXU18191.1 AF173372 Z46676 KQ460947 KPJ10531.1 ABLF02022069 ODYU01006284 SOQ47989.1 CH964232 EDW80737.1 CH379063 EDY72286.1 CM000162 EDX02121.1 CH480823 EDW56171.1 NEVH01020335 PNF21736.1 CH940650 EDW67401.2 CP012528 ALC49557.1 CVRI01000004 CRK87301.1 OUUW01000001 SPP73715.1 AE014298 AAF48976.1 KQ976441 KYM86395.1 AJVK01002698 AJVK01002699 AJVK01002700 GECL01000986 JAP05138.1 CM000364 EDX14073.1 GDHC01013884 JAQ04745.1 PCG71839.1 CM000366 EDX18400.1 CH479242 EDW37664.1 APCN01001143 LFJK02000006 POM47667.1 DS268408 EFO86322.1 GFTR01007282 JAW09144.1 GECU01014041 JAS93665.1 NNAY01003533 OXU19242.1 EDX02122.2 ADTU01003043 KQ977780 KYM99948.1 GL435766 EFN72620.1 KQ979074 KYN22900.1 AAAB01008846 EAU77249.3 CH916371 EDV92468.1 GALX01006776 JAB61690.1 PYGN01000947 PSN39086.1 GEBQ01007577 JAT32400.1 LIAE01010705 PAV56380.1 NMWX01000004 OZG03476.1 HE600919 CAP32259.2 CVRI01000044 CRK96891.1 KQ971338 EFA01831.2 KB632326 ERL92433.1 GL888262 EGI63691.1 NWSH01003464 PCG66235.1 PDUG01000002 PIC44143.1 GAMC01012119 JAB94436.1 ODYU01008317 SOQ51800.1 PNF21711.1 CM000160 EDW98157.2 GGMS01005383 MBY74586.1
Proteomes
UP000218220
UP000037510
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000005204 UP000019118 UP000007266 UP000007801 UP000002358 UP000245037 UP000215335 UP000001940 UP000192221 UP000007819 UP000095282 UP000007798 UP000001819 UP000075902 UP000002282 UP000001292 UP000235965 UP000008792 UP000092553 UP000183832 UP000075880 UP000268350 UP000000803 UP000078540 UP000092462 UP000000304 UP000008744 UP000005237 UP000075903 UP000075840 UP000076407 UP000237256 UP000008281 UP000005205 UP000078542 UP000000311 UP000078492 UP000007062 UP000001070 UP000218231 UP000216624 UP000075881 UP000008549 UP000030742 UP000007755 UP000230233
UP000005204 UP000019118 UP000007266 UP000007801 UP000002358 UP000245037 UP000215335 UP000001940 UP000192221 UP000007819 UP000095282 UP000007798 UP000001819 UP000075902 UP000002282 UP000001292 UP000235965 UP000008792 UP000092553 UP000183832 UP000075880 UP000268350 UP000000803 UP000078540 UP000092462 UP000000304 UP000008744 UP000005237 UP000075903 UP000075840 UP000076407 UP000237256 UP000008281 UP000005205 UP000078542 UP000000311 UP000078492 UP000007062 UP000001070 UP000218231 UP000216624 UP000075881 UP000008549 UP000030742 UP000007755 UP000230233
Interpro
ProteinModelPortal
A0A2A4IZN6
A0A2W1BP43
A0A0L7LIX9
A0A2H1VF27
A0A194RL37
A0A194PG52
+ More
A0A3S2LZ61 A0A212F1J8 H9J678 A0A2A4JXW1 A0A2H1VCF8 A0A1E1WQD5 A0A1E1WB37 A0A1E1W9B1 A0A1E1WIK4 A0A2H1VRN7 A0A2H1WD69 A0A0N0PE46 N6U8Y8 A0A194QEM0 A0A2H1W229 A0A2H1V1A7 D6WQE4 B3MQQ0 A0A1E1WGX5 A0A1E1WUR1 A0A2H1UZX8 A0A2A4JJD7 A0A1B6CQA8 A0A2A4JY64 A0A194PEK6 K7J4D4 A0A2P8YMN8 A0A232EIQ2 Q09225 A0A1W4VFZ1 A0A194R3W4 J9KT87 A0A2H1W4H1 A0A1I7UQZ1 B4NAC3 B5DL87 A0A182U2H9 B4PZG5 B4I773 A0A2J7PZI3 B4M0Z0 A0A0M4FAS0 A0A1J1HH25 A0A182JM14 A0A3B0IZN8 Q9VWG5 A0A195BMB6 A0A1B0D242 A0A0V0GAI0 B4R216 A0A146LAZ8 A0A2A4JJ41 B4R7R7 B4HAQ3 A0A2H2IGP4 A0A182V747 A0A182IAC3 A0A182XEF8 A0A2P4WDN8 E3LGZ1 A0A224XDS6 A0A1B6J389 A0A232ELQ7 B4PZG6 A0A158NXG1 A0A151IFH6 E2A1E7 A0A195ECF1 A0NCC8 B4JME3 V5FVL9 A0A2P8Y4A3 A0A1B6M8Z4 A0A2A2J3J9 A0A261B190 A0A182K8B2 A8XHZ3 A0A1J1I9K6 D1ZZM5 U4UJ36 F4WQ68 A0A2A4J2E4 A0A2G5UXB0 W8BM55 A0A2H1WFD8 K7J4D6 A0A2J7PZF1 B4PNU7 A0A2S2QA26
A0A3S2LZ61 A0A212F1J8 H9J678 A0A2A4JXW1 A0A2H1VCF8 A0A1E1WQD5 A0A1E1WB37 A0A1E1W9B1 A0A1E1WIK4 A0A2H1VRN7 A0A2H1WD69 A0A0N0PE46 N6U8Y8 A0A194QEM0 A0A2H1W229 A0A2H1V1A7 D6WQE4 B3MQQ0 A0A1E1WGX5 A0A1E1WUR1 A0A2H1UZX8 A0A2A4JJD7 A0A1B6CQA8 A0A2A4JY64 A0A194PEK6 K7J4D4 A0A2P8YMN8 A0A232EIQ2 Q09225 A0A1W4VFZ1 A0A194R3W4 J9KT87 A0A2H1W4H1 A0A1I7UQZ1 B4NAC3 B5DL87 A0A182U2H9 B4PZG5 B4I773 A0A2J7PZI3 B4M0Z0 A0A0M4FAS0 A0A1J1HH25 A0A182JM14 A0A3B0IZN8 Q9VWG5 A0A195BMB6 A0A1B0D242 A0A0V0GAI0 B4R216 A0A146LAZ8 A0A2A4JJ41 B4R7R7 B4HAQ3 A0A2H2IGP4 A0A182V747 A0A182IAC3 A0A182XEF8 A0A2P4WDN8 E3LGZ1 A0A224XDS6 A0A1B6J389 A0A232ELQ7 B4PZG6 A0A158NXG1 A0A151IFH6 E2A1E7 A0A195ECF1 A0NCC8 B4JME3 V5FVL9 A0A2P8Y4A3 A0A1B6M8Z4 A0A2A2J3J9 A0A261B190 A0A182K8B2 A8XHZ3 A0A1J1I9K6 D1ZZM5 U4UJ36 F4WQ68 A0A2A4J2E4 A0A2G5UXB0 W8BM55 A0A2H1WFD8 K7J4D6 A0A2J7PZF1 B4PNU7 A0A2S2QA26
Ontologies
GO
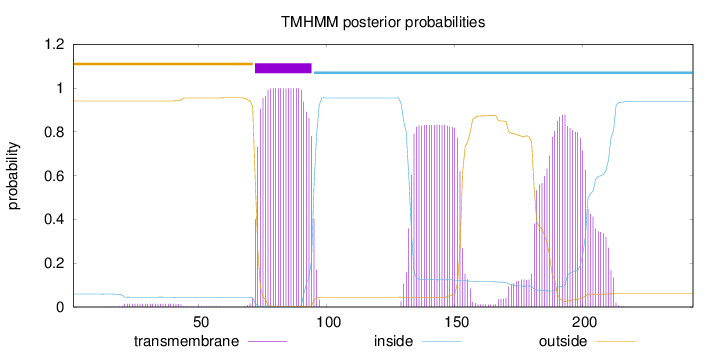
Topology
Subcellular location
Membrane
Length:
243
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
59.9821000000001
Exp number, first 60 AAs:
0.3615
Total prob of N-in:
0.05913
outside
1 - 71
TMhelix
72 - 94
inside
95 - 243
Population Genetic Test Statistics
Pi
345.057167
Theta
228.979879
Tajima's D
1.51906
CLR
0.221502
CSRT
0.790810459477026
Interpretation
Uncertain
