Gene
KWMTBOMO15246 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005015
Annotation
flavin-dependent_monooxygenase_FMO1_[Bombyx_mori]
Full name
Flavin-containing monooxygenase
+ More
Senecionine N-oxygenase
Dimethylaniline monooxygenase [N-oxide-forming]
Senecionine N-oxygenase
Dimethylaniline monooxygenase [N-oxide-forming]
Location in the cell
Cytoplasmic Reliability : 1.78
Sequence
CDS
ATGGAATATTACGACTTTCCATTTCCCGAGGGAACCCCGTCGTATCCATCGGCAACGTGCTTCCTCGATTACTTAAAATCGTTTGTGAAGCATTTCGATTTACTCAGTCATATTCAATTGCGCAGCTTGGTCACATCAGTGAAGTGGGCGGGCAATCACTGGAACCTTACTTACACAAAGACGGACACCAAGGAAAATGTAACTGAGACGTGTGACTTCATTGTGGTAGCAAATGGCCCTTATAATACTCCTGTGTGGCCCAAGTACGACGGAATAGACACATTCGAAGGCAGCATGATCCACAGCCATGATTACAAAGATCGGAAGGCGTACAAAAACAGAAAGGTGCTTATAGTAGGAGCTGGAGCTTCAGGCTTGGATCTCGCGATACAGTTGTCCAACGTGACCGCCAAATTAGTTCACAGCCACCACTTGGTGTATAACGAACCGAAATTTTTCGATGGTTACGTGAAGAAACCAGACATAATGGCGTTTACTCCTAAAGGCGTTATTTTCCGGGACGAATCATTTGAGGAATTGGATGATGTAATTTTTTGCACAGGTTATGACTTCAACCACCCGTTTCTAGACGAGAGCTGCGGTGTTACCTCAACCGCCAAATTTGTACTGCCTCTCCATAAGCAACTTGTCAATATAAAACATCCCAGCATGGTATTTCTTGGAATCGCCAAGAAGATTATAACAAGAGTTATGGATGCTCAGGCTGAATACGCAGCACTACTAGCTTCGGGTAAATTGAAGCTTCCATCACAGGAGGAAATGTTGAATTCTTGGCTGAAACACATCTCTTCCCTACAGGTTAAGGGCATGAAAATTATCGATTTAAACGTAGTCGGTTCGGAAATGGATCAATATTTCGGCAATTTAACTGAAGAAGCTGGTGTGGTGAGAGCGCCCCCGGTACTCACAGCGATCAGGGATTTCAACGGCGTCAACCGTTTGGATGATTTGCTCAACTACCGCGAATACGACTATAGCATTATTGATAATTTTCATTACGAAAGAAAATATAATCCAAGGCAGAACATACCTTGCCCTGTCGAAGTGTGA
Protein
MEYYDFPFPEGTPSYPSATCFLDYLKSFVKHFDLLSHIQLRSLVTSVKWAGNHWNLTYTKTDTKENVTETCDFIVVANGPYNTPVWPKYDGIDTFEGSMIHSHDYKDRKAYKNRKVLIVGAGASGLDLAIQLSNVTAKLVHSHHLVYNEPKFFDGYVKKPDIMAFTPKGVIFRDESFEELDDVIFCTGYDFNHPFLDESCGVTSTAKFVLPLHKQLVNIKHPSMVFLGIAKKIITRVMDAQAEYAALLASGKLKLPSQEEMLNSWLKHISSLQVKGMKIIDLNVVGSEMDQYFGNLTEEAGVVRAPPVLTAIRDFNGVNRLDDLLNYREYDYSIIDNFHYERKYNPRQNIPCPVEV
Summary
Description
NADPH-dependent monooxygenase that detoxifies senecionine and similar plant alkaloids that are ingested by the larvae. Is active towards a narrow range of related substrates with highest activity towards senecionine, followed by seneciphylline, retrorsine, monocrotaline, senecivernine, axillarine and axillaridine.
Catalytic Activity
NADPH + O2 + senecionine = H2O + NADP(+) + senecionine N-oxide
H(+) + N,N-dimethylaniline + NADPH + O2 = H2O + N,N-dimethylaniline N-oxide + NADP(+)
H(+) + N,N-dimethylaniline + NADPH + O2 = H2O + N,N-dimethylaniline N-oxide + NADP(+)
Cofactor
FAD
Biophysicochemical Properties
1.4 uM for senecionine
1.3 uM for NADPH
12.5 uM for monocrotaline
1.3 uM for NADPH
12.5 uM for monocrotaline
Subunit
Homotetramer.
Miscellaneous
Larvae store pyrrolizidine alkaloids from their host plant S.jacobaeae as non-toxic N-oxides. This serves as protection against insectivores; after ingestion of the larvae, the N-oxides are reduced in the gut of the insectivores to form toxic alkaloids (PubMed:11972041).
Similarity
Belongs to the FMO family.
Keywords
Direct protein sequencing
FAD
Flavoprotein
Monooxygenase
NADP
Oxidoreductase
Secreted
Signal
Feature
chain Flavin-containing monooxygenase
Uniprot
D7PGZ3
D7PGZ4
H9J674
A0A3S2LIM8
D7PGZ8
A0A2W1BRW5
+ More
A0A2A4JMJ1 A0A286R1L9 A0A194PGD2 A0A194RKS1 A0A212F1T7 A0A2H1VF51 D6CHF2 D6CHF1 D6CHF0 Q8MP06 A0A212FDL5 D6CHF3 A0A212FDS2 D6CHF7 A0A2H1VFD8 D7PGZ5 A0A286R1L3 D6CHF5 A0A194RQ38 D6CHF4 A0A0N1IQD4 D6CHF6 D6CHF8 A0A2W1BHH5 A0A0N1I9T0 D7PGZ9 A0A2A4JPB9 A0A2A4JPF0 D7PH00 A0A194PI12 H9J6M5 T1HWV6 R4G4T9 D7PGZ7 A0A212ERB5 S4P1Z8 A0A286R1N3 D7GXL5 D7PGZ6 A0A1B0CSN7 A0A2H8TMC0 A0A0N1IDD1 A0A1L8E4E9 D7PH01 A0A087ZTT5 A0A224XCP3 A0A084W6V0 J9JV88 A0A2A3EBU3 A0A154PME0 A0A1B0D7G2 A0A1L8E571 U5ETD5 A0A026WCD8 A0A182R8W8 A0A182QJN8 A0A2H1V3V2 A0A336MKL4 E2A9M6 A0A182PUE9 A0A194R599 A0A182YE75 A0A182IWA4 A0A0N0U3V8 A0A2M3Z5D9 A0A182SW70 Q17N35 A0A182FBL5 A0A1I8M338 T1PI45 W5JX01 A0A0K8WHR4 A0A182LSD8 A0A1S4EWV8 A0A146M349 A0A3B0JIE5 B4I8X8 W5JX83 A0A0A9Z036 A0A182NVD3 A0A2M4BPB0 A0A0A9Z1L3 A0A195BQ08 A0A0L0CIS3 A0A0C9R0F3 A0A034W0H1 A0A0J9U9S6 A0A2M4BPF2 N6TLD0 A0A2A4ITS9 U4UI72 A0A2M4BNW4 A0A2W1BRB6
A0A2A4JMJ1 A0A286R1L9 A0A194PGD2 A0A194RKS1 A0A212F1T7 A0A2H1VF51 D6CHF2 D6CHF1 D6CHF0 Q8MP06 A0A212FDL5 D6CHF3 A0A212FDS2 D6CHF7 A0A2H1VFD8 D7PGZ5 A0A286R1L3 D6CHF5 A0A194RQ38 D6CHF4 A0A0N1IQD4 D6CHF6 D6CHF8 A0A2W1BHH5 A0A0N1I9T0 D7PGZ9 A0A2A4JPB9 A0A2A4JPF0 D7PH00 A0A194PI12 H9J6M5 T1HWV6 R4G4T9 D7PGZ7 A0A212ERB5 S4P1Z8 A0A286R1N3 D7GXL5 D7PGZ6 A0A1B0CSN7 A0A2H8TMC0 A0A0N1IDD1 A0A1L8E4E9 D7PH01 A0A087ZTT5 A0A224XCP3 A0A084W6V0 J9JV88 A0A2A3EBU3 A0A154PME0 A0A1B0D7G2 A0A1L8E571 U5ETD5 A0A026WCD8 A0A182R8W8 A0A182QJN8 A0A2H1V3V2 A0A336MKL4 E2A9M6 A0A182PUE9 A0A194R599 A0A182YE75 A0A182IWA4 A0A0N0U3V8 A0A2M3Z5D9 A0A182SW70 Q17N35 A0A182FBL5 A0A1I8M338 T1PI45 W5JX01 A0A0K8WHR4 A0A182LSD8 A0A1S4EWV8 A0A146M349 A0A3B0JIE5 B4I8X8 W5JX83 A0A0A9Z036 A0A182NVD3 A0A2M4BPB0 A0A0A9Z1L3 A0A195BQ08 A0A0L0CIS3 A0A0C9R0F3 A0A034W0H1 A0A0J9U9S6 A0A2M4BPF2 N6TLD0 A0A2A4ITS9 U4UI72 A0A2M4BNW4 A0A2W1BRB6
EC Number
1.-.-.-
1.14.13.101
1.14.13.8
1.14.13.101
1.14.13.8
Pubmed
EMBL
GU564654
ADH16746.1
GU564655
ADH16747.1
BABH01019965
BABH01019966
+ More
BABH01019967 RSAL01000102 RVE47468.1 GU564659 ADH16751.1 KZ150046 PZC74503.1 NWSH01000977 PCG73295.1 MF465793 ASV48991.1 KQ459604 KPI92362.1 KQ460045 KPJ18137.1 AGBW02010802 OWR47700.1 ODYU01002253 SOQ39490.1 FN649424 CBI83748.1 FN649423 CBI83747.1 FN649422 CBI83746.1 AJ420233 AGBW02009018 OWR51849.1 FN649425 CBI83749.1 OWR51848.1 FN649429 CBI83753.1 SOQ39491.1 GU564656 ADH16748.1 MF465794 ASV48992.1 FN649427 CBI83751.1 KPJ18136.1 FN649426 CBI83750.1 LADI01010417 KPJ20713.1 FN649428 CBI83752.1 FN649430 CBI83754.1 PZC74502.1 KQ459383 KPJ01246.1 GU564660 ADH16752.1 PCG73293.1 PCG73292.1 GU564661 ADH16753.1 KPI92364.1 ACPB03023670 GAHY01000476 JAA77034.1 GU564658 ADH16750.1 AGBW02013066 OWR44027.1 GAIX01012245 JAA80315.1 MF465795 ASV48993.1 GG694257 EFA13493.1 BABH01041995 GU564657 ADH16749.1 AJWK01026328 GFXV01003498 MBW15303.1 KQ459169 KPJ03412.1 GFDF01000456 JAV13628.1 GU564662 ADH16754.1 GFTR01006259 JAW10167.1 ATLV01020959 KE525310 KFB45944.1 ABLF02031373 KZ288291 PBC29207.1 KQ434977 KZC12907.1 AJVK01003957 AJVK01003958 AJVK01003959 GFDF01000402 JAV13682.1 GANO01002830 JAB57041.1 KK107274 QOIP01000008 EZA53735.1 RLU19303.1 AXCN02002299 ODYU01000543 SOQ35479.1 UFQS01001190 UFQT01001190 SSX09607.1 SSX29403.1 GL437918 EFN69828.1 KQ460779 KPJ12415.1 KQ435851 KOX70828.1 GGFM01002990 MBW23741.1 CH477202 EAT48108.1 KA647573 AFP62202.1 ADMH02000071 ETN67910.1 GDHF01001623 JAI50691.1 AXCM01001621 GDHC01004411 JAQ14218.1 OUUW01000001 SPP73189.1 CH480824 EDW57053.1 ETN67909.1 GBHO01006368 GBHO01006367 GBRD01005428 GBRD01005427 GBRD01005425 GBRD01005423 JAG37236.1 JAG37237.1 JAG60393.1 GGFJ01005701 MBW54842.1 GBHO01006366 GBHO01006365 JAG37238.1 JAG37239.1 KQ976424 KYM88590.1 JRES01000340 KNC32117.1 GBYB01001475 JAG71242.1 GAKP01009862 GAKP01009861 JAC49091.1 CM002911 KMY96190.1 GGFJ01005743 MBW54884.1 APGK01019955 KB740150 ENN81259.1 NWSH01008153 PCG62674.1 KB632173 ERL89585.1 GGFJ01005629 MBW54770.1 KZ150001 PZC75310.1
BABH01019967 RSAL01000102 RVE47468.1 GU564659 ADH16751.1 KZ150046 PZC74503.1 NWSH01000977 PCG73295.1 MF465793 ASV48991.1 KQ459604 KPI92362.1 KQ460045 KPJ18137.1 AGBW02010802 OWR47700.1 ODYU01002253 SOQ39490.1 FN649424 CBI83748.1 FN649423 CBI83747.1 FN649422 CBI83746.1 AJ420233 AGBW02009018 OWR51849.1 FN649425 CBI83749.1 OWR51848.1 FN649429 CBI83753.1 SOQ39491.1 GU564656 ADH16748.1 MF465794 ASV48992.1 FN649427 CBI83751.1 KPJ18136.1 FN649426 CBI83750.1 LADI01010417 KPJ20713.1 FN649428 CBI83752.1 FN649430 CBI83754.1 PZC74502.1 KQ459383 KPJ01246.1 GU564660 ADH16752.1 PCG73293.1 PCG73292.1 GU564661 ADH16753.1 KPI92364.1 ACPB03023670 GAHY01000476 JAA77034.1 GU564658 ADH16750.1 AGBW02013066 OWR44027.1 GAIX01012245 JAA80315.1 MF465795 ASV48993.1 GG694257 EFA13493.1 BABH01041995 GU564657 ADH16749.1 AJWK01026328 GFXV01003498 MBW15303.1 KQ459169 KPJ03412.1 GFDF01000456 JAV13628.1 GU564662 ADH16754.1 GFTR01006259 JAW10167.1 ATLV01020959 KE525310 KFB45944.1 ABLF02031373 KZ288291 PBC29207.1 KQ434977 KZC12907.1 AJVK01003957 AJVK01003958 AJVK01003959 GFDF01000402 JAV13682.1 GANO01002830 JAB57041.1 KK107274 QOIP01000008 EZA53735.1 RLU19303.1 AXCN02002299 ODYU01000543 SOQ35479.1 UFQS01001190 UFQT01001190 SSX09607.1 SSX29403.1 GL437918 EFN69828.1 KQ460779 KPJ12415.1 KQ435851 KOX70828.1 GGFM01002990 MBW23741.1 CH477202 EAT48108.1 KA647573 AFP62202.1 ADMH02000071 ETN67910.1 GDHF01001623 JAI50691.1 AXCM01001621 GDHC01004411 JAQ14218.1 OUUW01000001 SPP73189.1 CH480824 EDW57053.1 ETN67909.1 GBHO01006368 GBHO01006367 GBRD01005428 GBRD01005427 GBRD01005425 GBRD01005423 JAG37236.1 JAG37237.1 JAG60393.1 GGFJ01005701 MBW54842.1 GBHO01006366 GBHO01006365 JAG37238.1 JAG37239.1 KQ976424 KYM88590.1 JRES01000340 KNC32117.1 GBYB01001475 JAG71242.1 GAKP01009862 GAKP01009861 JAC49091.1 CM002911 KMY96190.1 GGFJ01005743 MBW54884.1 APGK01019955 KB740150 ENN81259.1 NWSH01008153 PCG62674.1 KB632173 ERL89585.1 GGFJ01005629 MBW54770.1 KZ150001 PZC75310.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000015103 UP000007266 UP000092461 UP000005203 UP000030765 UP000007819 UP000242457 UP000076502 UP000092462 UP000053097 UP000279307 UP000075900 UP000075886 UP000000311 UP000075885 UP000076408 UP000075880 UP000053105 UP000075901 UP000008820 UP000069272 UP000095301 UP000000673 UP000075883 UP000268350 UP000001292 UP000075884 UP000078540 UP000037069 UP000019118 UP000030742
UP000015103 UP000007266 UP000092461 UP000005203 UP000030765 UP000007819 UP000242457 UP000076502 UP000092462 UP000053097 UP000279307 UP000075900 UP000075886 UP000000311 UP000075885 UP000076408 UP000075880 UP000053105 UP000075901 UP000008820 UP000069272 UP000095301 UP000000673 UP000075883 UP000268350 UP000001292 UP000075884 UP000078540 UP000037069 UP000019118 UP000030742
Pfam
PF00743 FMO-like
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
D7PGZ3
D7PGZ4
H9J674
A0A3S2LIM8
D7PGZ8
A0A2W1BRW5
+ More
A0A2A4JMJ1 A0A286R1L9 A0A194PGD2 A0A194RKS1 A0A212F1T7 A0A2H1VF51 D6CHF2 D6CHF1 D6CHF0 Q8MP06 A0A212FDL5 D6CHF3 A0A212FDS2 D6CHF7 A0A2H1VFD8 D7PGZ5 A0A286R1L3 D6CHF5 A0A194RQ38 D6CHF4 A0A0N1IQD4 D6CHF6 D6CHF8 A0A2W1BHH5 A0A0N1I9T0 D7PGZ9 A0A2A4JPB9 A0A2A4JPF0 D7PH00 A0A194PI12 H9J6M5 T1HWV6 R4G4T9 D7PGZ7 A0A212ERB5 S4P1Z8 A0A286R1N3 D7GXL5 D7PGZ6 A0A1B0CSN7 A0A2H8TMC0 A0A0N1IDD1 A0A1L8E4E9 D7PH01 A0A087ZTT5 A0A224XCP3 A0A084W6V0 J9JV88 A0A2A3EBU3 A0A154PME0 A0A1B0D7G2 A0A1L8E571 U5ETD5 A0A026WCD8 A0A182R8W8 A0A182QJN8 A0A2H1V3V2 A0A336MKL4 E2A9M6 A0A182PUE9 A0A194R599 A0A182YE75 A0A182IWA4 A0A0N0U3V8 A0A2M3Z5D9 A0A182SW70 Q17N35 A0A182FBL5 A0A1I8M338 T1PI45 W5JX01 A0A0K8WHR4 A0A182LSD8 A0A1S4EWV8 A0A146M349 A0A3B0JIE5 B4I8X8 W5JX83 A0A0A9Z036 A0A182NVD3 A0A2M4BPB0 A0A0A9Z1L3 A0A195BQ08 A0A0L0CIS3 A0A0C9R0F3 A0A034W0H1 A0A0J9U9S6 A0A2M4BPF2 N6TLD0 A0A2A4ITS9 U4UI72 A0A2M4BNW4 A0A2W1BRB6
A0A2A4JMJ1 A0A286R1L9 A0A194PGD2 A0A194RKS1 A0A212F1T7 A0A2H1VF51 D6CHF2 D6CHF1 D6CHF0 Q8MP06 A0A212FDL5 D6CHF3 A0A212FDS2 D6CHF7 A0A2H1VFD8 D7PGZ5 A0A286R1L3 D6CHF5 A0A194RQ38 D6CHF4 A0A0N1IQD4 D6CHF6 D6CHF8 A0A2W1BHH5 A0A0N1I9T0 D7PGZ9 A0A2A4JPB9 A0A2A4JPF0 D7PH00 A0A194PI12 H9J6M5 T1HWV6 R4G4T9 D7PGZ7 A0A212ERB5 S4P1Z8 A0A286R1N3 D7GXL5 D7PGZ6 A0A1B0CSN7 A0A2H8TMC0 A0A0N1IDD1 A0A1L8E4E9 D7PH01 A0A087ZTT5 A0A224XCP3 A0A084W6V0 J9JV88 A0A2A3EBU3 A0A154PME0 A0A1B0D7G2 A0A1L8E571 U5ETD5 A0A026WCD8 A0A182R8W8 A0A182QJN8 A0A2H1V3V2 A0A336MKL4 E2A9M6 A0A182PUE9 A0A194R599 A0A182YE75 A0A182IWA4 A0A0N0U3V8 A0A2M3Z5D9 A0A182SW70 Q17N35 A0A182FBL5 A0A1I8M338 T1PI45 W5JX01 A0A0K8WHR4 A0A182LSD8 A0A1S4EWV8 A0A146M349 A0A3B0JIE5 B4I8X8 W5JX83 A0A0A9Z036 A0A182NVD3 A0A2M4BPB0 A0A0A9Z1L3 A0A195BQ08 A0A0L0CIS3 A0A0C9R0F3 A0A034W0H1 A0A0J9U9S6 A0A2M4BPF2 N6TLD0 A0A2A4ITS9 U4UI72 A0A2M4BNW4 A0A2W1BRB6
PDB
5NMX
E-value=4.73285e-46,
Score=465
Ontologies
GO
Topology
Subcellular location
Secreted
Microsome membrane
Endoplasmic reticulum membrane
Microsome membrane
Endoplasmic reticulum membrane
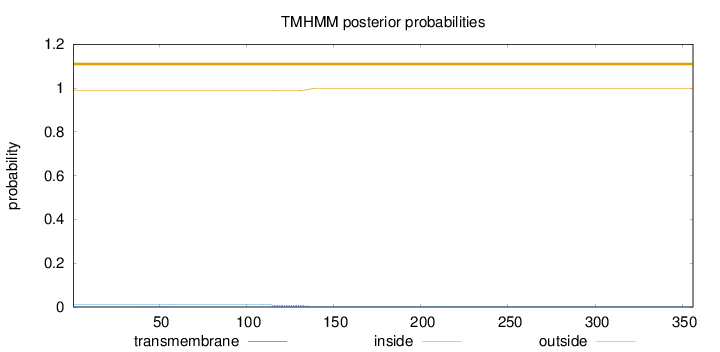
Length:
356
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17517
Exp number, first 60 AAs:
0.00411
Total prob of N-in:
0.01043
outside
1 - 356
Population Genetic Test Statistics
Pi
259.755149
Theta
162.254028
Tajima's D
1.842205
CLR
0.085805
CSRT
0.856407179641018
Interpretation
Uncertain
