Gene
KWMTBOMO15242
Pre Gene Modal
BGIBMGA005169
Annotation
transcription_initiation_factor_IIA_gamma_chain_[Bombyx_mori]
Full name
Transcription initiation factor IIA subunit 2
Alternative Name
General transcription factor IIA subunit 2
TFIIA p14 subunit
Transcription initiation factor IIA gamma chain
dTFIIA-S
TFIIA p14 subunit
Transcription initiation factor IIA gamma chain
dTFIIA-S
Location in the cell
Nuclear Reliability : 3.715
Sequence
CDS
ATGTCATATCAATTATACAGAAATACCACTATTGGTAATACACTTCAAGAAAGCCTAGACGAGCTGATCCAATATGGACAAATAACACCAGCACTAGCAGTAAAAGTTTTGCTGCAATTCGACAAATCTATAAATCAAGCTCTGTCCAATAGAGTAAAGTCTAGGCTAACCTTTAAAGCAGGAAAACTGAACACATACAGGTTTTGTGATAATGTATGGACTTTTATGTTGAATGATGTTGAATTTCGAGAAGTACAAGAATTGGCCAAGGTGGAAAAAGTTAAAATAGTAGCATGTGATGGTAAAAATGTTGACGACCGCAGATAA
Protein
MSYQLYRNTTIGNTLQESLDELIQYGQITPALAVKVLLQFDKSINQALSNRVKSRLTFKAGKLNTYRFCDNVWTFMLNDVEFREVQELAKVEKVKIVACDGKNVDDRR
Summary
Description
TFIIA is a component of the transcription machinery of RNA polymerase II and plays an important role in transcriptional activation.
TFIIA is a component of the transcription machinery of RNA polymerase II and plays an important role in transcriptional activation (PubMed:7958898). TFIIA in a complex with TBP mediates transcriptional activity (PubMed:7958898). Part of a rhi-dependent transcription machinery that enables the generation of piRNA precursors from heterochromatin while maintaining the suppression of transposon-encoded promoters and enhancers (PubMed:28847004). Forms a complex with Moonshiner/CG12721 and Trf2 which recruit transcriptional machinery to heterochromatin to initiate the bidirectional transcription of piRNA clusters, by interacting with the RDC (rhi, del and cuff) complex that binds to repressive H3K9me3 marks in the chromatin (PubMed:28847004). This mechanism allows transcription to occur in piRNA clusters despite the lack of proper promoter elements and in the presence of the repressive H3K9me3 mark (PubMed:28847004).
TFIIA is a component of the transcription machinery of RNA polymerase II and plays an important role in transcriptional activation (PubMed:7958898). TFIIA in a complex with TBP mediates transcriptional activity (PubMed:7958898). Part of a rhi-dependent transcription machinery that enables the generation of piRNA precursors from heterochromatin while maintaining the suppression of transposon-encoded promoters and enhancers (PubMed:28847004). Forms a complex with Moonshiner/CG12721 and Trf2 which recruit transcriptional machinery to heterochromatin to initiate the bidirectional transcription of piRNA clusters, by interacting with the RDC (rhi, del and cuff) complex that binds to repressive H3K9me3 marks in the chromatin (PubMed:28847004). This mechanism allows transcription to occur in piRNA clusters despite the lack of proper promoter elements and in the presence of the repressive H3K9me3 mark (PubMed:28847004).
Subunit
TFIIA is a heterodimer of the large unprocessed subunit 1 and a small subunit gamma (PubMed:7958898). It was originally believed to be a heterotrimer of an alpha (p30), a beta (p20) and a gamma (p14) subunit (PubMed:7958898). Forms a complex with Moonshiner/CG12721 and Trf2 (PubMed:28847004).
Similarity
Belongs to the TFIIA subunit 2 family.
Keywords
Complete proteome
Direct protein sequencing
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain Transcription initiation factor IIA subunit 2
Uniprot
Q1HPQ3
A0A2W1BMK6
A0A2H1VLL1
A0A2A4JNA2
A0A1E1WL83
A0A212F1T3
+ More
S4PTT6 H9J6M7 A0A194RQ43 A0A194PH33 A0A1B6CI72 A0A026W3N7 V5FWB1 A0A195CMA0 A0A195CRM4 F4WS91 V9IKI4 A0A087ZWY8 A0A158NIM0 A0A310SRI4 A0A151XDU4 E1ZWR8 A0A1B6LJW3 A0A232ERF6 A0A2J7R2U2 A0A1W4WRZ3 A0A1B6FLB4 A0A1B6I0Y3 A0A2P8ZF93 A0A023EEX3 A0A0L7RG16 K7IUJ4 T1E3A2 Q17J10 A0A154P6E8 U5EK68 A0A1Y1N5J6 A0A182QLP1 J3JUB6 E2BFC2 A0A1Q3FWM1 A0A0T6B231 Q17J11 A0A2A3EU00 A0A182PLE1 A0A182KG68 A0A182XF36 Q7QA87 A2I432 D6WU86 A0A0A9Y581 A0A1B0ESI8 A0A0J7KWF4 A0A182W351 A0A182LZU4 A0A182RI55 A0A182SN83 A0A1L8DGN8 B0WJG2 B4M071 A0A1S4EQY7 A0A182HVK4 A0A2H8TGQ2 C4WSL4 B4KDS5 B4N825 A0A2S2R874 B4JTB0 A0A1B2AK92 P52656 A0A0M3QYG7 Q299B6 B4G4Z7 A0A1W4U8J5 B4R261 A0A3B0JL68 B4PNZ0 B4HFZ6 B3P8D1 A0A1J1J259 A0A023FAP4 A0A224XXI6 A0A0V0GBQ3 A0A0P4VKG8 R4FMM2 B3LXP8 A0A1B0G018 A0A1A9XVL4 A0A1A9VGD9 A0A1B0BKZ1 A0A1A9ZWW1 A0A1A9WNH4 E0VX10 T1P8P6 A0A2S2P363 A0A336L2C9 A0A023GH03 G3MSJ9 A0A224Z1M2 A0A131YVL8
S4PTT6 H9J6M7 A0A194RQ43 A0A194PH33 A0A1B6CI72 A0A026W3N7 V5FWB1 A0A195CMA0 A0A195CRM4 F4WS91 V9IKI4 A0A087ZWY8 A0A158NIM0 A0A310SRI4 A0A151XDU4 E1ZWR8 A0A1B6LJW3 A0A232ERF6 A0A2J7R2U2 A0A1W4WRZ3 A0A1B6FLB4 A0A1B6I0Y3 A0A2P8ZF93 A0A023EEX3 A0A0L7RG16 K7IUJ4 T1E3A2 Q17J10 A0A154P6E8 U5EK68 A0A1Y1N5J6 A0A182QLP1 J3JUB6 E2BFC2 A0A1Q3FWM1 A0A0T6B231 Q17J11 A0A2A3EU00 A0A182PLE1 A0A182KG68 A0A182XF36 Q7QA87 A2I432 D6WU86 A0A0A9Y581 A0A1B0ESI8 A0A0J7KWF4 A0A182W351 A0A182LZU4 A0A182RI55 A0A182SN83 A0A1L8DGN8 B0WJG2 B4M071 A0A1S4EQY7 A0A182HVK4 A0A2H8TGQ2 C4WSL4 B4KDS5 B4N825 A0A2S2R874 B4JTB0 A0A1B2AK92 P52656 A0A0M3QYG7 Q299B6 B4G4Z7 A0A1W4U8J5 B4R261 A0A3B0JL68 B4PNZ0 B4HFZ6 B3P8D1 A0A1J1J259 A0A023FAP4 A0A224XXI6 A0A0V0GBQ3 A0A0P4VKG8 R4FMM2 B3LXP8 A0A1B0G018 A0A1A9XVL4 A0A1A9VGD9 A0A1B0BKZ1 A0A1A9ZWW1 A0A1A9WNH4 E0VX10 T1P8P6 A0A2S2P363 A0A336L2C9 A0A023GH03 G3MSJ9 A0A224Z1M2 A0A131YVL8
Pubmed
28756777
22118469
23622113
19121390
26354079
24508170
+ More
30249741 21719571 21347285 20798317 28648823 29403074 24945155 26483478 20075255 24330624 17510324 28004739 22516182 23537049 12364791 14747013 17210077 18362917 19820115 25401762 26823975 17994087 7958898 10731132 12537572 12537569 28847004 15632085 17550304 25474469 27129103 20566863 25315136 22216098 28797301 26830274
30249741 21719571 21347285 20798317 28648823 29403074 24945155 26483478 20075255 24330624 17510324 28004739 22516182 23537049 12364791 14747013 17210077 18362917 19820115 25401762 26823975 17994087 7958898 10731132 12537572 12537569 28847004 15632085 17550304 25474469 27129103 20566863 25315136 22216098 28797301 26830274
EMBL
DQ443349
ABF51438.1
KZ150046
PZC74507.1
ODYU01003230
SOQ41720.1
+ More
NWSH01000977 PCG73299.1 GDQN01003292 JAT87762.1 AGBW02010802 OWR47697.1 GAIX01011473 JAA81087.1 BABH01019974 KQ460045 KPJ18141.1 KQ459604 KPI92358.1 GEDC01024159 JAS13139.1 KK107503 QOIP01000006 EZA49659.1 RLU22101.1 GALX01006531 JAB61935.1 KQ977634 KYN01209.1 KQ977372 KYN03285.1 GL888307 EGI62927.1 JR052162 AEY61633.1 ADTU01016949 KQ759888 OAD62119.1 KQ982254 KYQ58542.1 GL434901 EFN74357.1 GEBQ01015974 JAT24003.1 NNAY01002610 OXU20953.1 NEVH01007825 PNF35158.1 GECZ01018787 JAS50982.1 GECU01027099 JAS80607.1 PYGN01000075 PSN55171.1 JXUM01135077 GAPW01006194 GAPW01006193 GAPW01006192 GAPW01006191 KQ568218 JAC07406.1 KXJ69091.1 KQ414597 KOC69887.1 AAZX01002625 GALA01000024 JAA94828.1 CH477235 EAT46701.1 KQ434827 KZC07516.1 GANO01001925 JAB57946.1 GEZM01014336 JAV92140.1 AXCN02000338 APGK01028609 BT126829 KB740694 KB631600 AEE61791.1 ENN79448.1 ERL84526.1 GL448009 EFN85582.1 GFDL01003179 JAV31866.1 LJIG01016137 KRT81503.1 EAT46700.1 KZ288189 PBC34619.1 AAAB01008898 EAA09250.4 EF070535 ABM55601.1 KQ971352 EFA06253.1 GBHO01016808 GBRD01007295 GDHC01000929 JAG26796.1 JAG58526.1 JAQ17700.1 AJWK01004372 LBMM01002408 KMQ94867.1 AXCM01000063 GFDF01008580 JAV05504.1 DS231959 EDS29136.1 CH940650 EDW68321.2 APCN01005796 GFXV01000653 MBW12458.1 ABLF02038565 AK340239 BAH70884.1 CH933806 EDW14922.1 CH964232 EDW81276.1 GGMS01016920 MBY86123.1 CH916373 EDV95000.1 KX531771 ANY27581.1 X83271 AE014297 AY113478 BT044069 CP012526 ALC47589.1 CM000070 EAL27787.2 CH479179 EDW24663.1 CM000364 EDX14118.1 OUUW01000007 SPP83064.1 CM000160 EDW98200.1 CH480815 EDW43389.1 CH954182 EDV53955.1 CVRI01000066 CRL05852.1 GBBI01000603 JAC18109.1 GFTR01001778 JAW14648.1 GECL01001351 JAP04773.1 GDKW01001023 JAI55572.1 ACPB03031360 GAHY01000928 JAA76582.1 CH902617 EDV43942.1 CCAG010000726 JXJN01016126 DS235824 EEB17916.1 KA644490 AFP59119.1 GGMR01011251 MBY23870.1 UFQS01001369 UFQT01001369 SSX10543.1 SSX30229.1 GBBM01003150 JAC32268.1 JO844850 AEO36467.1 GFPF01012602 MAA23748.1 GEDV01006351 JAP82206.1
NWSH01000977 PCG73299.1 GDQN01003292 JAT87762.1 AGBW02010802 OWR47697.1 GAIX01011473 JAA81087.1 BABH01019974 KQ460045 KPJ18141.1 KQ459604 KPI92358.1 GEDC01024159 JAS13139.1 KK107503 QOIP01000006 EZA49659.1 RLU22101.1 GALX01006531 JAB61935.1 KQ977634 KYN01209.1 KQ977372 KYN03285.1 GL888307 EGI62927.1 JR052162 AEY61633.1 ADTU01016949 KQ759888 OAD62119.1 KQ982254 KYQ58542.1 GL434901 EFN74357.1 GEBQ01015974 JAT24003.1 NNAY01002610 OXU20953.1 NEVH01007825 PNF35158.1 GECZ01018787 JAS50982.1 GECU01027099 JAS80607.1 PYGN01000075 PSN55171.1 JXUM01135077 GAPW01006194 GAPW01006193 GAPW01006192 GAPW01006191 KQ568218 JAC07406.1 KXJ69091.1 KQ414597 KOC69887.1 AAZX01002625 GALA01000024 JAA94828.1 CH477235 EAT46701.1 KQ434827 KZC07516.1 GANO01001925 JAB57946.1 GEZM01014336 JAV92140.1 AXCN02000338 APGK01028609 BT126829 KB740694 KB631600 AEE61791.1 ENN79448.1 ERL84526.1 GL448009 EFN85582.1 GFDL01003179 JAV31866.1 LJIG01016137 KRT81503.1 EAT46700.1 KZ288189 PBC34619.1 AAAB01008898 EAA09250.4 EF070535 ABM55601.1 KQ971352 EFA06253.1 GBHO01016808 GBRD01007295 GDHC01000929 JAG26796.1 JAG58526.1 JAQ17700.1 AJWK01004372 LBMM01002408 KMQ94867.1 AXCM01000063 GFDF01008580 JAV05504.1 DS231959 EDS29136.1 CH940650 EDW68321.2 APCN01005796 GFXV01000653 MBW12458.1 ABLF02038565 AK340239 BAH70884.1 CH933806 EDW14922.1 CH964232 EDW81276.1 GGMS01016920 MBY86123.1 CH916373 EDV95000.1 KX531771 ANY27581.1 X83271 AE014297 AY113478 BT044069 CP012526 ALC47589.1 CM000070 EAL27787.2 CH479179 EDW24663.1 CM000364 EDX14118.1 OUUW01000007 SPP83064.1 CM000160 EDW98200.1 CH480815 EDW43389.1 CH954182 EDV53955.1 CVRI01000066 CRL05852.1 GBBI01000603 JAC18109.1 GFTR01001778 JAW14648.1 GECL01001351 JAP04773.1 GDKW01001023 JAI55572.1 ACPB03031360 GAHY01000928 JAA76582.1 CH902617 EDV43942.1 CCAG010000726 JXJN01016126 DS235824 EEB17916.1 KA644490 AFP59119.1 GGMR01011251 MBY23870.1 UFQS01001369 UFQT01001369 SSX10543.1 SSX30229.1 GBBM01003150 JAC32268.1 JO844850 AEO36467.1 GFPF01012602 MAA23748.1 GEDV01006351 JAP82206.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000053240
UP000053268
UP000053097
+ More
UP000279307 UP000078542 UP000007755 UP000005203 UP000005205 UP000075809 UP000000311 UP000215335 UP000235965 UP000192223 UP000245037 UP000069940 UP000249989 UP000053825 UP000002358 UP000008820 UP000076502 UP000075886 UP000019118 UP000030742 UP000008237 UP000242457 UP000075885 UP000075881 UP000076407 UP000007062 UP000007266 UP000092461 UP000036403 UP000075920 UP000075883 UP000075900 UP000075901 UP000002320 UP000008792 UP000079169 UP000075840 UP000007819 UP000009192 UP000007798 UP000001070 UP000000803 UP000092553 UP000001819 UP000008744 UP000192221 UP000000304 UP000268350 UP000002282 UP000001292 UP000008711 UP000183832 UP000015103 UP000007801 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000009046 UP000095301
UP000279307 UP000078542 UP000007755 UP000005203 UP000005205 UP000075809 UP000000311 UP000215335 UP000235965 UP000192223 UP000245037 UP000069940 UP000249989 UP000053825 UP000002358 UP000008820 UP000076502 UP000075886 UP000019118 UP000030742 UP000008237 UP000242457 UP000075885 UP000075881 UP000076407 UP000007062 UP000007266 UP000092461 UP000036403 UP000075920 UP000075883 UP000075900 UP000075901 UP000002320 UP000008792 UP000079169 UP000075840 UP000007819 UP000009192 UP000007798 UP000001070 UP000000803 UP000092553 UP000001819 UP000008744 UP000192221 UP000000304 UP000268350 UP000002282 UP000001292 UP000008711 UP000183832 UP000015103 UP000007801 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000009046 UP000095301
Interpro
SUPFAM
SSF50784
SSF50784
Gene 3D
ProteinModelPortal
Q1HPQ3
A0A2W1BMK6
A0A2H1VLL1
A0A2A4JNA2
A0A1E1WL83
A0A212F1T3
+ More
S4PTT6 H9J6M7 A0A194RQ43 A0A194PH33 A0A1B6CI72 A0A026W3N7 V5FWB1 A0A195CMA0 A0A195CRM4 F4WS91 V9IKI4 A0A087ZWY8 A0A158NIM0 A0A310SRI4 A0A151XDU4 E1ZWR8 A0A1B6LJW3 A0A232ERF6 A0A2J7R2U2 A0A1W4WRZ3 A0A1B6FLB4 A0A1B6I0Y3 A0A2P8ZF93 A0A023EEX3 A0A0L7RG16 K7IUJ4 T1E3A2 Q17J10 A0A154P6E8 U5EK68 A0A1Y1N5J6 A0A182QLP1 J3JUB6 E2BFC2 A0A1Q3FWM1 A0A0T6B231 Q17J11 A0A2A3EU00 A0A182PLE1 A0A182KG68 A0A182XF36 Q7QA87 A2I432 D6WU86 A0A0A9Y581 A0A1B0ESI8 A0A0J7KWF4 A0A182W351 A0A182LZU4 A0A182RI55 A0A182SN83 A0A1L8DGN8 B0WJG2 B4M071 A0A1S4EQY7 A0A182HVK4 A0A2H8TGQ2 C4WSL4 B4KDS5 B4N825 A0A2S2R874 B4JTB0 A0A1B2AK92 P52656 A0A0M3QYG7 Q299B6 B4G4Z7 A0A1W4U8J5 B4R261 A0A3B0JL68 B4PNZ0 B4HFZ6 B3P8D1 A0A1J1J259 A0A023FAP4 A0A224XXI6 A0A0V0GBQ3 A0A0P4VKG8 R4FMM2 B3LXP8 A0A1B0G018 A0A1A9XVL4 A0A1A9VGD9 A0A1B0BKZ1 A0A1A9ZWW1 A0A1A9WNH4 E0VX10 T1P8P6 A0A2S2P363 A0A336L2C9 A0A023GH03 G3MSJ9 A0A224Z1M2 A0A131YVL8
S4PTT6 H9J6M7 A0A194RQ43 A0A194PH33 A0A1B6CI72 A0A026W3N7 V5FWB1 A0A195CMA0 A0A195CRM4 F4WS91 V9IKI4 A0A087ZWY8 A0A158NIM0 A0A310SRI4 A0A151XDU4 E1ZWR8 A0A1B6LJW3 A0A232ERF6 A0A2J7R2U2 A0A1W4WRZ3 A0A1B6FLB4 A0A1B6I0Y3 A0A2P8ZF93 A0A023EEX3 A0A0L7RG16 K7IUJ4 T1E3A2 Q17J10 A0A154P6E8 U5EK68 A0A1Y1N5J6 A0A182QLP1 J3JUB6 E2BFC2 A0A1Q3FWM1 A0A0T6B231 Q17J11 A0A2A3EU00 A0A182PLE1 A0A182KG68 A0A182XF36 Q7QA87 A2I432 D6WU86 A0A0A9Y581 A0A1B0ESI8 A0A0J7KWF4 A0A182W351 A0A182LZU4 A0A182RI55 A0A182SN83 A0A1L8DGN8 B0WJG2 B4M071 A0A1S4EQY7 A0A182HVK4 A0A2H8TGQ2 C4WSL4 B4KDS5 B4N825 A0A2S2R874 B4JTB0 A0A1B2AK92 P52656 A0A0M3QYG7 Q299B6 B4G4Z7 A0A1W4U8J5 B4R261 A0A3B0JL68 B4PNZ0 B4HFZ6 B3P8D1 A0A1J1J259 A0A023FAP4 A0A224XXI6 A0A0V0GBQ3 A0A0P4VKG8 R4FMM2 B3LXP8 A0A1B0G018 A0A1A9XVL4 A0A1A9VGD9 A0A1B0BKZ1 A0A1A9ZWW1 A0A1A9WNH4 E0VX10 T1P8P6 A0A2S2P363 A0A336L2C9 A0A023GH03 G3MSJ9 A0A224Z1M2 A0A131YVL8
PDB
6O9L
E-value=1.42535e-37,
Score=385
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
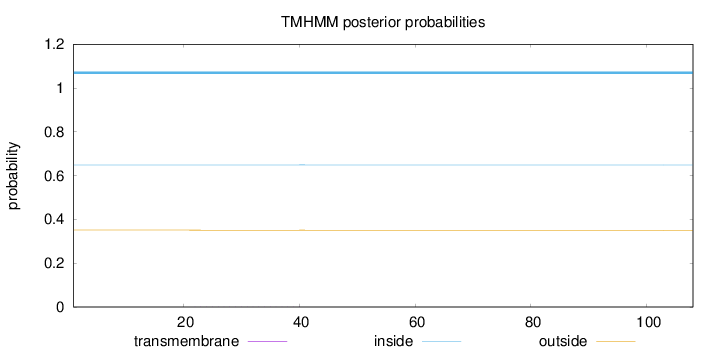
Length:
108
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00953
Exp number, first 60 AAs:
0.00883
Total prob of N-in:
0.64880
inside
1 - 108
Population Genetic Test Statistics
Pi
200.265781
Theta
187.312152
Tajima's D
0.798508
CLR
0.005517
CSRT
0.595870206489675
Interpretation
Uncertain
