Gene
KWMTBOMO15234 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005010
Annotation
PREDICTED:_ubiquinone_biosynthesis_monooxygenase_COQ6?_mitochondrial_[Bombyx_mori]
Full name
Ubiquinone biosynthesis monooxygenase COQ6, mitochondrial
Alternative Name
Coenzyme Q10 monooxygenase 6
Location in the cell
Mitochondrial Reliability : 1.634
Sequence
CDS
ATGGGTGTGGTGGCCACCTTGTACTTAGCCGAGGAAACGGATAACACAACTGCCTGGCAAAGATGGCTGCCCTCAGGTCCAATAGCCATCCTGCCTTTGGACTCGCAGCGTAGCTCACTCGTCTGGTCGACGTGGGAGGACAACGCGAAACGACTCTTGAAACTACCCGAGGAACAATTCGTCGATGAACTCAACGATGCCCTTTGGAAGCAGTATCCTCGGAGCGGCGCAGTGGAGGGCTGCATGTCGTGGGTGAGCGGCTGCCTGCGTCGGGCCGGACTACCGGACGGTGCGGCGCGCCAGCTCCCGCCTTCCGTGCAAGGAGTAGCGTCCTCCTCCCGCGCCGCTTTCCCCCTCGCCTTCGGGCACAGTACACGATACGTCGCTAACGGAGTAGCTCTAGTCGGAGATGCGGCCCACAGAGTCCATCCGCTGGCTGGACAGGGCGTTAACCTCGGATTTGGCGACGTAAAGGAACTCACGGAACTGCTAGCCGACGCCACATACGGCGGCTTCGATATTAACGATCCGAGCTACCTGCAGAGATACGAGAGTAACCGTCAGAGGCATAACGTGCCGACTCAGCTCGCTATAGAGGCCCTCCACCGGCTCTACACGGTCGACCTGCCTCCGGTCGTGCTGCTGAGGAGTGTCGGCCTCCAACTGACAAACGCTCTACATCCTGTCAAGAAATTATTAATGTCTCACGCTGCAACGTAG
Protein
MGVVATLYLAEETDNTTAWQRWLPSGPIAILPLDSQRSSLVWSTWEDNAKRLLKLPEEQFVDELNDALWKQYPRSGAVEGCMSWVSGCLRRAGLPDGAARQLPPSVQGVASSSRAAFPLAFGHSTRYVANGVALVGDAAHRVHPLAGQGVNLGFGDVKELTELLADATYGGFDINDPSYLQRYESNRQRHNVPTQLAIEALHRLYTVDLPPVVLLRSVGLQLTNALHPVKKLLMSHAAT
Summary
Description
FAD-dependent monooxygenase required for the C5-ring hydroxylation during ubiquinone biosynthesis. Catalyzes the hydroxylation of 3-polyprenyl-4-hydroxybenzoic acid to 3-polyprenyl-4,5-dihydroxybenzoic acid. The electrons required for the hydroxylation reaction may be funneled indirectly from NADPH via a ferredoxin/ferredoxin reductase system to COQ6.
Cofactor
FAD
Subunit
Component of a multi-subunit COQ enzyme complex.
Component of a multi-subunit COQ enzyme complex, composed of at least COQ3, COQ4, COQ5, COQ6, COQ7 and COQ9. Interacts with ADCK4 and COQ7.
Component of a multi-subunit COQ enzyme complex, composed of at least coq3, coq4, coq5, coq6, coq7 and coq9. Interacts with coq8b and coq7.
Component of a multi-subunit COQ enzyme complex, composed of at least COQ3, COQ4, COQ5, COQ6, COQ7 and COQ9. Interacts with ADCK4 and COQ7.
Component of a multi-subunit COQ enzyme complex, composed of at least coq3, coq4, coq5, coq6, coq7 and coq9. Interacts with coq8b and coq7.
Similarity
Belongs to the UbiH/COQ6 family.
Keywords
Cell projection
Complete proteome
FAD
Flavoprotein
Golgi apparatus
Membrane
Mitochondrion
Mitochondrion inner membrane
Monooxygenase
Oxidoreductase
Reference proteome
Transit peptide
Ubiquinone biosynthesis
Feature
chain Ubiquinone biosynthesis monooxygenase COQ6, mitochondrial
Uniprot
H9J669
A0A3S2LZA8
A0A194PGC2
A0A2A4J127
A0A2W1BIY2
S4PSV2
+ More
A0A2H1W860 A0A194RLF2 K7JED2 A0A026WIN1 A0A3L8D7B0 A0A232ESL8 A0A2J7Q225 A0A151WR83 A0A154P3J3 A0A195BY98 A0A088A361 A0A195EG56 A0A158NI89 A0A195AWK6 A0A0N0U755 A0A2A3E8X6 E2C8K8 A0A0C9Q6W0 U5EJF9 A0A0C9R8M6 A0A0C9R127 J3JWE3 A0A182GUL0 A0A1Y1KQA6 A0A067QHI4 A0A2L2YHY1 Q1DGZ9 A0A310SI00 E2APB4 A0A195FEZ9 A0A182HIK4 A0A139WF39 A0A182KY78 A0A182JWI7 A0A182PFC4 A0A182R6D3 W4Y3Y9 A0A182STQ3 A0A182W410 E0W075 W5MYN5 Q0IFQ5 A0A182M1J0 A0A212FEZ1 A0A2C9H898 F1RAX8 A0A034W8T8 A0A182UCM3 V9KLF4 A0A0A1X366 A0A182UN08 A0A1Q3G445 A0A087T878 R7V989 W8BQT3 C3XS22 A0A151NKA4 S4RFM1 S4RFM3 A0A2R8Q6S9 A0A182IW11 B4JAF7 A0A0L0BZ71 B0WV09 K7G9A7 Q7QER0 A0A182YQW7 B4LU17 A0A2H6NMH8 A0A3Q0GC79 A0A2D0RRD1 A0A3B4E2Q0 T1P9T1 A0A1I8MBS7 I3KWA8 V8NPZ8 H3DJ31 A0A1J1J3R1 Q6DF46 W4YG09 A0A091IBM0 A0A1L8F969 G3UUV4 Q4QQX3 A0A3B5A032 A0A1L8F0Q6 F1NBR7 E9JB46 A0A3B4ZX06 M4AL43 T1DCP6 A0A218VEU9 A0A093R5Y9
A0A2H1W860 A0A194RLF2 K7JED2 A0A026WIN1 A0A3L8D7B0 A0A232ESL8 A0A2J7Q225 A0A151WR83 A0A154P3J3 A0A195BY98 A0A088A361 A0A195EG56 A0A158NI89 A0A195AWK6 A0A0N0U755 A0A2A3E8X6 E2C8K8 A0A0C9Q6W0 U5EJF9 A0A0C9R8M6 A0A0C9R127 J3JWE3 A0A182GUL0 A0A1Y1KQA6 A0A067QHI4 A0A2L2YHY1 Q1DGZ9 A0A310SI00 E2APB4 A0A195FEZ9 A0A182HIK4 A0A139WF39 A0A182KY78 A0A182JWI7 A0A182PFC4 A0A182R6D3 W4Y3Y9 A0A182STQ3 A0A182W410 E0W075 W5MYN5 Q0IFQ5 A0A182M1J0 A0A212FEZ1 A0A2C9H898 F1RAX8 A0A034W8T8 A0A182UCM3 V9KLF4 A0A0A1X366 A0A182UN08 A0A1Q3G445 A0A087T878 R7V989 W8BQT3 C3XS22 A0A151NKA4 S4RFM1 S4RFM3 A0A2R8Q6S9 A0A182IW11 B4JAF7 A0A0L0BZ71 B0WV09 K7G9A7 Q7QER0 A0A182YQW7 B4LU17 A0A2H6NMH8 A0A3Q0GC79 A0A2D0RRD1 A0A3B4E2Q0 T1P9T1 A0A1I8MBS7 I3KWA8 V8NPZ8 H3DJ31 A0A1J1J3R1 Q6DF46 W4YG09 A0A091IBM0 A0A1L8F969 G3UUV4 Q4QQX3 A0A3B5A032 A0A1L8F0Q6 F1NBR7 E9JB46 A0A3B4ZX06 M4AL43 T1DCP6 A0A218VEU9 A0A093R5Y9
EC Number
1.14.13.-
Pubmed
19121390
26354079
28756777
23622113
20075255
24508170
+ More
30249741 28648823 21347285 20798317 22516182 26483478 28004739 24845553 26561354 17510324 18362917 19820115 20966253 20566863 22118469 23594743 25348373 24402279 25830018 23254933 24495485 18563158 22293439 17994087 26108605 17381049 12364791 14747013 17210077 25244985 25315136 25186727 24297900 15496914 20431018 27762356 20838655 15592404 21282665 23542700 23758969
30249741 28648823 21347285 20798317 22516182 26483478 28004739 24845553 26561354 17510324 18362917 19820115 20966253 20566863 22118469 23594743 25348373 24402279 25830018 23254933 24495485 18563158 22293439 17994087 26108605 17381049 12364791 14747013 17210077 25244985 25315136 25186727 24297900 15496914 20431018 27762356 20838655 15592404 21282665 23542700 23758969
EMBL
BABH01019983
BABH01019984
RSAL01000102
RVE47474.1
KQ459604
KPI92352.1
+ More
NWSH01004460 PCG65083.1 KZ150014 PZC75019.1 GAIX01013143 JAA79417.1 ODYU01006958 SOQ49279.1 KQ460045 KPJ18145.1 AAZX01007490 KK107185 EZA55833.1 QOIP01000012 RLU16365.1 NNAY01002421 OXU21307.1 NEVH01019373 PNF22636.1 KQ982813 KYQ50311.1 KQ434809 KZC06392.1 KQ978501 KYM93567.1 KQ978957 KYN27223.1 ADTU01016099 ADTU01016100 KQ976725 KYM76556.1 KQ435719 KOX78758.1 KZ288322 PBC28175.1 GL453666 EFN75738.1 GBYB01009843 JAG79610.1 GANO01002249 JAB57622.1 GBYB01012719 JAG82486.1 GBYB01009844 JAG79611.1 BT127561 AEE62523.1 JXUM01089336 KQ563763 KXJ73353.1 GEZM01079905 GEZM01079904 JAV62350.1 KK853628 KDR03814.1 IAAA01021616 LAA07743.1 CH899941 EAT32429.1 KQ761039 OAD58343.1 GL441504 EFN64713.1 KQ981636 KYN38797.1 APCN01003992 KQ971353 KYB26590.1 AAGJ04124569 AAGJ04124570 AAZO01006645 DS235857 EEB19031.1 AHAT01003744 CH477294 EAT44481.1 AXCM01004049 AGBW02008891 OWR52290.1 BX936443 BC122260 GAKP01007828 JAC51124.1 JW866370 AFO98887.1 GBXI01008720 JAD05572.1 GFDL01000474 JAV34571.1 KK113905 KFM61317.1 AMQN01000789 KB295623 ELU12926.1 GAMC01005133 JAC01423.1 GG666456 EEN69490.1 AKHW03002792 KYO37217.1 CH916368 EDW03828.1 JRES01001119 KNC25362.1 DS232115 EDS35337.1 AGCU01050015 AGCU01050016 AAAB01008846 EAA06342.5 CH940649 EDW65070.1 IACI01120289 IACI01120290 LAA35763.1 KA645511 AFP60140.1 AERX01030277 AZIM01002372 ETE64145.1 CVRI01000069 CRL07024.1 CR761032 AAMC01082860 BC076897 KL217612 KFO97160.1 CM004480 OCT68139.1 BC097912 AAH97912.1 CM004481 OCT65145.1 AADN05000303 GL770938 EFZ09924.1 GAAZ01000574 JAA97369.1 MUZQ01000005 OWK64102.1 KL430757 KFW91567.1
NWSH01004460 PCG65083.1 KZ150014 PZC75019.1 GAIX01013143 JAA79417.1 ODYU01006958 SOQ49279.1 KQ460045 KPJ18145.1 AAZX01007490 KK107185 EZA55833.1 QOIP01000012 RLU16365.1 NNAY01002421 OXU21307.1 NEVH01019373 PNF22636.1 KQ982813 KYQ50311.1 KQ434809 KZC06392.1 KQ978501 KYM93567.1 KQ978957 KYN27223.1 ADTU01016099 ADTU01016100 KQ976725 KYM76556.1 KQ435719 KOX78758.1 KZ288322 PBC28175.1 GL453666 EFN75738.1 GBYB01009843 JAG79610.1 GANO01002249 JAB57622.1 GBYB01012719 JAG82486.1 GBYB01009844 JAG79611.1 BT127561 AEE62523.1 JXUM01089336 KQ563763 KXJ73353.1 GEZM01079905 GEZM01079904 JAV62350.1 KK853628 KDR03814.1 IAAA01021616 LAA07743.1 CH899941 EAT32429.1 KQ761039 OAD58343.1 GL441504 EFN64713.1 KQ981636 KYN38797.1 APCN01003992 KQ971353 KYB26590.1 AAGJ04124569 AAGJ04124570 AAZO01006645 DS235857 EEB19031.1 AHAT01003744 CH477294 EAT44481.1 AXCM01004049 AGBW02008891 OWR52290.1 BX936443 BC122260 GAKP01007828 JAC51124.1 JW866370 AFO98887.1 GBXI01008720 JAD05572.1 GFDL01000474 JAV34571.1 KK113905 KFM61317.1 AMQN01000789 KB295623 ELU12926.1 GAMC01005133 JAC01423.1 GG666456 EEN69490.1 AKHW03002792 KYO37217.1 CH916368 EDW03828.1 JRES01001119 KNC25362.1 DS232115 EDS35337.1 AGCU01050015 AGCU01050016 AAAB01008846 EAA06342.5 CH940649 EDW65070.1 IACI01120289 IACI01120290 LAA35763.1 KA645511 AFP60140.1 AERX01030277 AZIM01002372 ETE64145.1 CVRI01000069 CRL07024.1 CR761032 AAMC01082860 BC076897 KL217612 KFO97160.1 CM004480 OCT68139.1 BC097912 AAH97912.1 CM004481 OCT65145.1 AADN05000303 GL770938 EFZ09924.1 GAAZ01000574 JAA97369.1 MUZQ01000005 OWK64102.1 KL430757 KFW91567.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000053240
UP000002358
+ More
UP000053097 UP000279307 UP000215335 UP000235965 UP000075809 UP000076502 UP000078542 UP000005203 UP000078492 UP000005205 UP000078540 UP000053105 UP000242457 UP000008237 UP000069940 UP000249989 UP000027135 UP000008820 UP000000311 UP000078541 UP000075840 UP000007266 UP000075882 UP000075881 UP000075885 UP000075900 UP000007110 UP000075901 UP000075920 UP000009046 UP000018468 UP000075883 UP000007151 UP000076407 UP000000437 UP000075902 UP000075903 UP000054359 UP000014760 UP000001554 UP000050525 UP000245300 UP000075880 UP000001070 UP000037069 UP000002320 UP000007267 UP000007062 UP000076408 UP000008792 UP000189705 UP000221080 UP000261440 UP000095301 UP000005207 UP000007303 UP000183832 UP000008143 UP000054308 UP000186698 UP000001645 UP000261400 UP000000539 UP000002852 UP000197619
UP000053097 UP000279307 UP000215335 UP000235965 UP000075809 UP000076502 UP000078542 UP000005203 UP000078492 UP000005205 UP000078540 UP000053105 UP000242457 UP000008237 UP000069940 UP000249989 UP000027135 UP000008820 UP000000311 UP000078541 UP000075840 UP000007266 UP000075882 UP000075881 UP000075885 UP000075900 UP000007110 UP000075901 UP000075920 UP000009046 UP000018468 UP000075883 UP000007151 UP000076407 UP000000437 UP000075902 UP000075903 UP000054359 UP000014760 UP000001554 UP000050525 UP000245300 UP000075880 UP000001070 UP000037069 UP000002320 UP000007267 UP000007062 UP000076408 UP000008792 UP000189705 UP000221080 UP000261440 UP000095301 UP000005207 UP000007303 UP000183832 UP000008143 UP000054308 UP000186698 UP000001645 UP000261400 UP000000539 UP000002852 UP000197619
Interpro
Gene 3D
ProteinModelPortal
H9J669
A0A3S2LZA8
A0A194PGC2
A0A2A4J127
A0A2W1BIY2
S4PSV2
+ More
A0A2H1W860 A0A194RLF2 K7JED2 A0A026WIN1 A0A3L8D7B0 A0A232ESL8 A0A2J7Q225 A0A151WR83 A0A154P3J3 A0A195BY98 A0A088A361 A0A195EG56 A0A158NI89 A0A195AWK6 A0A0N0U755 A0A2A3E8X6 E2C8K8 A0A0C9Q6W0 U5EJF9 A0A0C9R8M6 A0A0C9R127 J3JWE3 A0A182GUL0 A0A1Y1KQA6 A0A067QHI4 A0A2L2YHY1 Q1DGZ9 A0A310SI00 E2APB4 A0A195FEZ9 A0A182HIK4 A0A139WF39 A0A182KY78 A0A182JWI7 A0A182PFC4 A0A182R6D3 W4Y3Y9 A0A182STQ3 A0A182W410 E0W075 W5MYN5 Q0IFQ5 A0A182M1J0 A0A212FEZ1 A0A2C9H898 F1RAX8 A0A034W8T8 A0A182UCM3 V9KLF4 A0A0A1X366 A0A182UN08 A0A1Q3G445 A0A087T878 R7V989 W8BQT3 C3XS22 A0A151NKA4 S4RFM1 S4RFM3 A0A2R8Q6S9 A0A182IW11 B4JAF7 A0A0L0BZ71 B0WV09 K7G9A7 Q7QER0 A0A182YQW7 B4LU17 A0A2H6NMH8 A0A3Q0GC79 A0A2D0RRD1 A0A3B4E2Q0 T1P9T1 A0A1I8MBS7 I3KWA8 V8NPZ8 H3DJ31 A0A1J1J3R1 Q6DF46 W4YG09 A0A091IBM0 A0A1L8F969 G3UUV4 Q4QQX3 A0A3B5A032 A0A1L8F0Q6 F1NBR7 E9JB46 A0A3B4ZX06 M4AL43 T1DCP6 A0A218VEU9 A0A093R5Y9
A0A2H1W860 A0A194RLF2 K7JED2 A0A026WIN1 A0A3L8D7B0 A0A232ESL8 A0A2J7Q225 A0A151WR83 A0A154P3J3 A0A195BY98 A0A088A361 A0A195EG56 A0A158NI89 A0A195AWK6 A0A0N0U755 A0A2A3E8X6 E2C8K8 A0A0C9Q6W0 U5EJF9 A0A0C9R8M6 A0A0C9R127 J3JWE3 A0A182GUL0 A0A1Y1KQA6 A0A067QHI4 A0A2L2YHY1 Q1DGZ9 A0A310SI00 E2APB4 A0A195FEZ9 A0A182HIK4 A0A139WF39 A0A182KY78 A0A182JWI7 A0A182PFC4 A0A182R6D3 W4Y3Y9 A0A182STQ3 A0A182W410 E0W075 W5MYN5 Q0IFQ5 A0A182M1J0 A0A212FEZ1 A0A2C9H898 F1RAX8 A0A034W8T8 A0A182UCM3 V9KLF4 A0A0A1X366 A0A182UN08 A0A1Q3G445 A0A087T878 R7V989 W8BQT3 C3XS22 A0A151NKA4 S4RFM1 S4RFM3 A0A2R8Q6S9 A0A182IW11 B4JAF7 A0A0L0BZ71 B0WV09 K7G9A7 Q7QER0 A0A182YQW7 B4LU17 A0A2H6NMH8 A0A3Q0GC79 A0A2D0RRD1 A0A3B4E2Q0 T1P9T1 A0A1I8MBS7 I3KWA8 V8NPZ8 H3DJ31 A0A1J1J3R1 Q6DF46 W4YG09 A0A091IBM0 A0A1L8F969 G3UUV4 Q4QQX3 A0A3B5A032 A0A1L8F0Q6 F1NBR7 E9JB46 A0A3B4ZX06 M4AL43 T1DCP6 A0A218VEU9 A0A093R5Y9
PDB
4K22
E-value=1.04594e-16,
Score=209
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion inner membrane
Golgi apparatus Localizes to cell processes and Golgi apparatus in podocytes. With evidence from 1 publications.
Cell projection Localizes to cell processes and Golgi apparatus in podocytes. With evidence from 1 publications.
Golgi apparatus Localizes to cell processes and Golgi apparatus in podocytes. With evidence from 1 publications.
Cell projection Localizes to cell processes and Golgi apparatus in podocytes. With evidence from 1 publications.
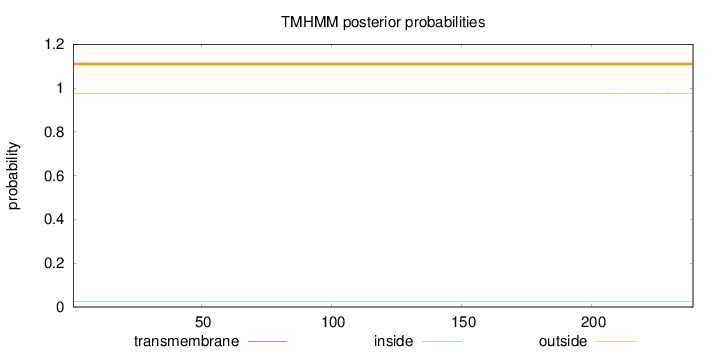
Length:
239
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00741
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.02533
outside
1 - 239
Population Genetic Test Statistics
Pi
4.94979
Theta
204.624869
Tajima's D
2.391667
CLR
0.967113
CSRT
0.934903254837258
Interpretation
Uncertain
