Gene
KWMTBOMO15233
Pre Gene Modal
BGIBMGA005170
Annotation
transient_receptor_potential_channel_pyrexia_[Bombyx_mori]
Full name
Transient receptor potential channel pyrexia
Location in the cell
Cytoplasmic Reliability : 3.063
Sequence
CDS
ATGAGTCAATTGCCGGAGGATGTAGAGGATGATGTCGAAGGAGGATACATATCTAGCGACGAGTCAGCACACGGCGCTGATCTCGCTGAGCGTCTGAGGGTCAAACCTTCTAAAGATATATGGGAACTGGACGAGATACAGTACACGTTACGGCTACTGCCGTATTCAGAAGAAATATCAGTACTTATAGGTAATGGGAATTACGATGAAACAATACAGTTTGCCTCCGAGGCTGAAGGCGGATTTGGCCTACGAACAGCCATCCTTTGGGCCTGTTGGCTCGGAAAGAGTATTCTGCTTTTTAAATTGCTAAAAATGGGCGTTGATCCCGATGAACTAGACGATGCCGGAAGGACATGTTTACATTTAAGTTGTTTAGTTGGAAGCGAAGAATGCGTGAAGCTTCTACTCGACCACGGAGCGCACCCGAATACATGGGACTCGTCTACAGAAACAAAAGCGACGCCTCTCCATTGCGCAGCCAGCGCCAAAAGTTTAGCTTGCGTAAAGGTGTTGATCGCTCATGGTGCTGACGTAAATGCCGGTTTAAGTGACAGGTCTCCGTTACATTACGCCGTCCTCAGCGATGCACCGGAAGTTGTGAAGGAATTATTAGAAGCTGGTGCGTGCCCCGACACGCCACAGGTGGGCTGTCTAAATTCATTTCAAACACAAACCAGATACTAA
Protein
MSQLPEDVEDDVEGGYISSDESAHGADLAERLRVKPSKDIWELDEIQYTLRLLPYSEEISVLIGNGNYDETIQFASEAEGGFGLRTAILWACWLGKSILLFKLLKMGVDPDELDDAGRTCLHLSCLVGSEECVKLLLDHGAHPNTWDSSTETKATPLHCAASAKSLACVKVLIAHGADVNAGLSDRSPLHYAVLSDAPEVVKELLEAGACPDTPQVGCLNSFQTQTRY
Summary
Description
Receptor-activated non-selective cation channel involved in protection or tolerance from high temperature stress. Activated by temperatures above 40 degrees Celsius. More permeable to K(+) than to Na(+). May act in stress protection allow flies to survive in natural environments.
Subunit
Homooligomer; between isoform A and isoform B.
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Belongs to the transient receptor (TC 1.A.4) family. STrpC subfamily.
Belongs to the transient receptor (TC 1.A.4) family. STrpC subfamily.
Keywords
Alternative splicing
ANK repeat
Complete proteome
Glycoprotein
Ion channel
Ion transport
Membrane
Reference proteome
Repeat
Sensory transduction
Transmembrane
Transmembrane helix
Transport
Feature
chain Transient receptor potential channel pyrexia
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J6M8
W8VMI8
A0A0L7K8K9
A0A3S2NSL9
A0A2W1BQK8
A0A2A4IZ93
+ More
A0A2A4J0Z3 A0A2H1VL68 A0A194RQ48 A0A194PG37 A0A212FEW5 A0A1Y1LNQ8 A0A067QSF9 A0A2J7Q265 D6WP79 A0A2P8YP14 B0WV08 A0A182GUK9 W5JS40 A0A182FJL5 A0A182W409 A0A182JWI8 Q0IFQ4 A0A182YQW8 A0A182T252 A0A182MX57 A0A182R6D4 A0A182Q024 E0W076 A0A1W4WPQ0 A0A182HIK5 Q7QEQ9 A0A182UYH1 A0A182WY66 A0A1J1J7Z5 A0A1A9WB91 A0A182JF36 B3M8Z5 A0A182N044 A0A1B0BMT9 A0A0M4EKT2 A0A1A9XD69 A0A336M1Y6 A0A336MEW4 B4H5D3 Q29ER3 A0A336MAH4 A0A1B0A764 B4KKC1 A0A3B0JY03 A0A1B0FHA3 B4QKU8 B4HVD0 A0A0J9UBA1 B4LS35 A0A1I8NXE6 B3NEM1 A0A1I8NAZ6 Q9W0T5 A0A1A9UH95 B4PC33 W8BAK3 A0A0L0BVP7 W8BFJ6 B4JQN9 B4N427 A0A182PFC5 A0A0K2SXU5
A0A2A4J0Z3 A0A2H1VL68 A0A194RQ48 A0A194PG37 A0A212FEW5 A0A1Y1LNQ8 A0A067QSF9 A0A2J7Q265 D6WP79 A0A2P8YP14 B0WV08 A0A182GUK9 W5JS40 A0A182FJL5 A0A182W409 A0A182JWI8 Q0IFQ4 A0A182YQW8 A0A182T252 A0A182MX57 A0A182R6D4 A0A182Q024 E0W076 A0A1W4WPQ0 A0A182HIK5 Q7QEQ9 A0A182UYH1 A0A182WY66 A0A1J1J7Z5 A0A1A9WB91 A0A182JF36 B3M8Z5 A0A182N044 A0A1B0BMT9 A0A0M4EKT2 A0A1A9XD69 A0A336M1Y6 A0A336MEW4 B4H5D3 Q29ER3 A0A336MAH4 A0A1B0A764 B4KKC1 A0A3B0JY03 A0A1B0FHA3 B4QKU8 B4HVD0 A0A0J9UBA1 B4LS35 A0A1I8NXE6 B3NEM1 A0A1I8NAZ6 Q9W0T5 A0A1A9UH95 B4PC33 W8BAK3 A0A0L0BVP7 W8BFJ6 B4JQN9 B4N427 A0A182PFC5 A0A0K2SXU5
Pubmed
EMBL
BABH01019984
BABH01019985
AB437368
BAO53209.1
JTDY01010234
KOB59632.1
+ More
RSAL01000102 RVE47475.1 KZ150014 PZC75020.1 NWSH01004460 PCG65081.1 PCG65082.1 ODYU01003165 SOQ41565.1 KQ460045 KPJ18146.1 KQ459604 KPI92351.1 AGBW02008891 OWR52289.1 GEZM01050962 GEZM01050961 JAV75259.1 KK853628 KDR03815.1 NEVH01019373 PNF22682.1 KQ971353 EFA07512.1 PYGN01000457 PSN45985.1 DS232115 EDS35336.1 JXUM01089330 JXUM01089331 KQ563763 KXJ73352.1 ADMH02000338 ETN66911.1 CH477294 EAT44480.1 AXCM01004049 AXCN02002023 AAZO01006645 DS235857 EEB19032.1 APCN01003993 AAAB01008846 EAA06365.5 CVRI01000069 CRL07025.1 CH902618 EDV38939.1 JXJN01017053 CP012525 ALC43922.1 UFQS01000411 UFQT01000411 SSX03695.1 SSX24060.1 UFQS01000805 UFQT01000805 SSX06999.1 SSX27343.1 CH479211 EDW32985.1 CH379070 EAL29997.2 SSX06998.1 SSX27342.1 CH933807 EDW12652.1 OUUW01000012 SPP86934.1 CCAG010019361 CM000363 EDX08694.1 CH480817 EDW49895.1 CM002912 KMY96600.1 CH940649 EDW64721.1 KRF81817.1 CH954178 EDV50016.1 AE014296 BT001363 BT010317 CM000159 EDW92688.1 GAMC01010838 JAB95717.1 JRES01001267 KNC24063.1 GAMC01010837 JAB95718.1 CH916372 EDV99219.1 CH964095 EDW78901.1 HACA01001213 CDW18574.1
RSAL01000102 RVE47475.1 KZ150014 PZC75020.1 NWSH01004460 PCG65081.1 PCG65082.1 ODYU01003165 SOQ41565.1 KQ460045 KPJ18146.1 KQ459604 KPI92351.1 AGBW02008891 OWR52289.1 GEZM01050962 GEZM01050961 JAV75259.1 KK853628 KDR03815.1 NEVH01019373 PNF22682.1 KQ971353 EFA07512.1 PYGN01000457 PSN45985.1 DS232115 EDS35336.1 JXUM01089330 JXUM01089331 KQ563763 KXJ73352.1 ADMH02000338 ETN66911.1 CH477294 EAT44480.1 AXCM01004049 AXCN02002023 AAZO01006645 DS235857 EEB19032.1 APCN01003993 AAAB01008846 EAA06365.5 CVRI01000069 CRL07025.1 CH902618 EDV38939.1 JXJN01017053 CP012525 ALC43922.1 UFQS01000411 UFQT01000411 SSX03695.1 SSX24060.1 UFQS01000805 UFQT01000805 SSX06999.1 SSX27343.1 CH479211 EDW32985.1 CH379070 EAL29997.2 SSX06998.1 SSX27342.1 CH933807 EDW12652.1 OUUW01000012 SPP86934.1 CCAG010019361 CM000363 EDX08694.1 CH480817 EDW49895.1 CM002912 KMY96600.1 CH940649 EDW64721.1 KRF81817.1 CH954178 EDV50016.1 AE014296 BT001363 BT010317 CM000159 EDW92688.1 GAMC01010838 JAB95717.1 JRES01001267 KNC24063.1 GAMC01010837 JAB95718.1 CH916372 EDV99219.1 CH964095 EDW78901.1 HACA01001213 CDW18574.1
Proteomes
UP000005204
UP000037510
UP000283053
UP000218220
UP000053240
UP000053268
+ More
UP000007151 UP000027135 UP000235965 UP000007266 UP000245037 UP000002320 UP000069940 UP000249989 UP000000673 UP000069272 UP000075920 UP000075881 UP000008820 UP000076408 UP000075901 UP000075883 UP000075900 UP000075886 UP000009046 UP000192223 UP000075840 UP000007062 UP000075903 UP000076407 UP000183832 UP000091820 UP000075880 UP000007801 UP000075884 UP000092460 UP000092553 UP000092443 UP000008744 UP000001819 UP000092445 UP000009192 UP000268350 UP000092444 UP000000304 UP000001292 UP000008792 UP000095300 UP000008711 UP000095301 UP000000803 UP000078200 UP000002282 UP000037069 UP000001070 UP000007798 UP000075885
UP000007151 UP000027135 UP000235965 UP000007266 UP000245037 UP000002320 UP000069940 UP000249989 UP000000673 UP000069272 UP000075920 UP000075881 UP000008820 UP000076408 UP000075901 UP000075883 UP000075900 UP000075886 UP000009046 UP000192223 UP000075840 UP000007062 UP000075903 UP000076407 UP000183832 UP000091820 UP000075880 UP000007801 UP000075884 UP000092460 UP000092553 UP000092443 UP000008744 UP000001819 UP000092445 UP000009192 UP000268350 UP000092444 UP000000304 UP000001292 UP000008792 UP000095300 UP000008711 UP000095301 UP000000803 UP000078200 UP000002282 UP000037069 UP000001070 UP000007798 UP000075885
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
H9J6M8
W8VMI8
A0A0L7K8K9
A0A3S2NSL9
A0A2W1BQK8
A0A2A4IZ93
+ More
A0A2A4J0Z3 A0A2H1VL68 A0A194RQ48 A0A194PG37 A0A212FEW5 A0A1Y1LNQ8 A0A067QSF9 A0A2J7Q265 D6WP79 A0A2P8YP14 B0WV08 A0A182GUK9 W5JS40 A0A182FJL5 A0A182W409 A0A182JWI8 Q0IFQ4 A0A182YQW8 A0A182T252 A0A182MX57 A0A182R6D4 A0A182Q024 E0W076 A0A1W4WPQ0 A0A182HIK5 Q7QEQ9 A0A182UYH1 A0A182WY66 A0A1J1J7Z5 A0A1A9WB91 A0A182JF36 B3M8Z5 A0A182N044 A0A1B0BMT9 A0A0M4EKT2 A0A1A9XD69 A0A336M1Y6 A0A336MEW4 B4H5D3 Q29ER3 A0A336MAH4 A0A1B0A764 B4KKC1 A0A3B0JY03 A0A1B0FHA3 B4QKU8 B4HVD0 A0A0J9UBA1 B4LS35 A0A1I8NXE6 B3NEM1 A0A1I8NAZ6 Q9W0T5 A0A1A9UH95 B4PC33 W8BAK3 A0A0L0BVP7 W8BFJ6 B4JQN9 B4N427 A0A182PFC5 A0A0K2SXU5
A0A2A4J0Z3 A0A2H1VL68 A0A194RQ48 A0A194PG37 A0A212FEW5 A0A1Y1LNQ8 A0A067QSF9 A0A2J7Q265 D6WP79 A0A2P8YP14 B0WV08 A0A182GUK9 W5JS40 A0A182FJL5 A0A182W409 A0A182JWI8 Q0IFQ4 A0A182YQW8 A0A182T252 A0A182MX57 A0A182R6D4 A0A182Q024 E0W076 A0A1W4WPQ0 A0A182HIK5 Q7QEQ9 A0A182UYH1 A0A182WY66 A0A1J1J7Z5 A0A1A9WB91 A0A182JF36 B3M8Z5 A0A182N044 A0A1B0BMT9 A0A0M4EKT2 A0A1A9XD69 A0A336M1Y6 A0A336MEW4 B4H5D3 Q29ER3 A0A336MAH4 A0A1B0A764 B4KKC1 A0A3B0JY03 A0A1B0FHA3 B4QKU8 B4HVD0 A0A0J9UBA1 B4LS35 A0A1I8NXE6 B3NEM1 A0A1I8NAZ6 Q9W0T5 A0A1A9UH95 B4PC33 W8BAK3 A0A0L0BVP7 W8BFJ6 B4JQN9 B4N427 A0A182PFC5 A0A0K2SXU5
PDB
5EIL
E-value=5.62851e-17,
Score=211
Ontologies
GO
Topology
Subcellular location
Membrane
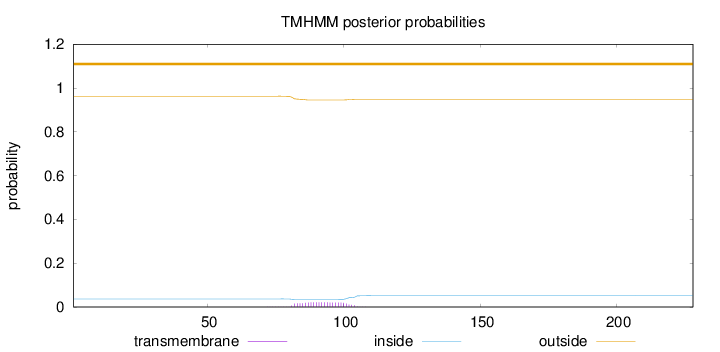
Length:
228
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43233
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.03694
outside
1 - 228
Population Genetic Test Statistics
Pi
183.470451
Theta
164.765545
Tajima's D
0.329465
CLR
0.684989
CSRT
0.466526673666317
Interpretation
Uncertain
