Gene
KWMTBOMO15231
Pre Gene Modal
BGIBMGA005009
Annotation
putative_ubiquinone_biosynthesis_monooxygenase_COQ6?_partial_[Operophtera_brumata]
Full name
Ubiquinone biosynthesis monooxygenase COQ6, mitochondrial
Alternative Name
Coenzyme Q10 monooxygenase 6
Location in the cell
Extracellular Reliability : 1.995
Sequence
CDS
ATGGTTGGCTGCACCTTGGCTTGTGCCTTAGCTAAAAACTCACTATTTTCCACTTTCAAAATATTACTATTGGAAAGCAGCCCTTACAAGCAATTTGAACTGAGAAAAGAATACAGCAATAGAGTTGTGGCACTTAGTCAAAATACCAAATCATTAATGTCATCTTTGAATGTTTGGAAACATATTGAAGATATGAGACTGCAACCTGTAAAACATATGCAAGTTTGGGATGCTTGTTCAGATGCTTTAATATCATTCAACAGTGCCGACAACTACAATGATGATGTTGCCTACATTGTCGAAAATGATGTATTGCTGCACGCTGTCAATACAGAACTAAATAATACAGACAATAAAAACTTTAATATTGTCTATGAAGCAAAGATTGCAGATTATACACTGCCAAATTCTGAAGAAATAAATCCAAAGAGTTTAGTTAAAATGAACAATGGTGACACATATTCATGCCAACTGCTGATTGAAATTAGTATGTTGGATTAG
Protein
MVGCTLACALAKNSLFSTFKILLLESSPYKQFELRKEYSNRVVALSQNTKSLMSSLNVWKHIEDMRLQPVKHMQVWDACSDALISFNSADNYNDDVAYIVENDVLLHAVNTELNNTDNKNFNIVYEAKIADYTLPNSEEINPKSLVKMNNGDTYSCQLLIEISMLD
Summary
Description
FAD-dependent monooxygenase required for the C5-ring hydroxylation during ubiquinone biosynthesis. Catalyzes the hydroxylation of 3-polyprenyl-4-hydroxybenzoic acid to 3-polyprenyl-4,5-dihydroxybenzoic acid. The electrons required for the hydroxylation reaction may be funneled indirectly from NADPH via a ferredoxin/ferredoxin reductase system to COQ6.
Cofactor
FAD
Subunit
Component of a multi-subunit COQ enzyme complex.
Component of a multi-subunit COQ enzyme complex, composed of at least COQ3, COQ4, COQ5, COQ6, COQ7 and COQ9. Interacts with ADCK4 and COQ7.
Component of a multi-subunit COQ enzyme complex, composed of at least COQ3, COQ4, COQ5, COQ6, COQ7 and COQ9. Interacts with ADCK4 and COQ7.
Similarity
Belongs to the UbiH/COQ6 family.
Keywords
Complete proteome
FAD
Flavoprotein
Membrane
Mitochondrion
Mitochondrion inner membrane
Monooxygenase
Oxidoreductase
Reference proteome
Transit peptide
Ubiquinone biosynthesis
Feature
chain Ubiquinone biosynthesis monooxygenase COQ6, mitochondrial
Uniprot
A0A0L7KQN6
A0A2H1VL88
A0A2W1BIY2
A0A2A4IZL9
A0A194RLF2
A0A212FEZ2
+ More
S4P958 A0A194PMK8 A0A026WIJ2 K7JED1 A0A232ESL8 V9IIU2 A0A2A3E8X6 E2C8K8 A0A088A361 A0A154P3J3 A0A3L8D6P3 A0A0N0U755 A0A158NI87 A0A310SI00 A0A195FFJ8 A0A195AWD4 A0A195BY98 A0A0C9R127 A0A1S3IS81 A0A151WR83 A0A0L7QXE3 A0A195EGE9 A0A139WFD9 A0A182FJL6 E2APB4 W5JRB1 A0A182N043 B4JAF7 A0A067QHI4 A0A182R6D5 A0A0P4WEC7 A0A2M3Z7R3 A0A2M3YXK0 A0A182MID6 A0A2M4AN04 A0A2M4BMD5 A0A2M4BMG8 A0A2M4CMM9 E0W075 A0A2M4CV75 A0A182JWI9 V4CJ86 A0A182U5L8 A0A182HIK6 Q0IFQ5 A0A182VIR1 A0A0L8GWX2 A0A2M4CWL9 A0A182TCB2 A0A1I8MBS7 V9KLF4 B4KF33 A0A084WFL1 T1P9T1 A0A182YQW7 B4LU17 A0A182GUK8 A0A1W4UIH1 Q7QER0 A0A2C9H898 A0A182QHV1 A0A1B6EMM7 A0A336KKH4 B4MWC8 A0A1Q3G445 H2VBJ8 U5EJF9 A0A023ETR5 A0A3P9LXR2 A0A3Q2EKU4 H3DJ31 A0A3P9KCV8 Q4RLY4 A0A3Q3VJP4 A0A2J7Q269 A0A3Q1ECR3 A0A3Q1GWD2 A0A1Y1KQA6 B5X2C0 A0A1B6KZM5 A0A3B5JZF5 A0A1S2ZMU0 A0A1I8Q2L8 A0A182PFC6 A0A3Q3ESC1 A0A2D0RRD1 A0A3Q1C556 A0A3Q0RFW6 B3N4W9 Q9VMQ5 A0A1B0FH47 D3TNK7 A0A1A9UQZ6
S4P958 A0A194PMK8 A0A026WIJ2 K7JED1 A0A232ESL8 V9IIU2 A0A2A3E8X6 E2C8K8 A0A088A361 A0A154P3J3 A0A3L8D6P3 A0A0N0U755 A0A158NI87 A0A310SI00 A0A195FFJ8 A0A195AWD4 A0A195BY98 A0A0C9R127 A0A1S3IS81 A0A151WR83 A0A0L7QXE3 A0A195EGE9 A0A139WFD9 A0A182FJL6 E2APB4 W5JRB1 A0A182N043 B4JAF7 A0A067QHI4 A0A182R6D5 A0A0P4WEC7 A0A2M3Z7R3 A0A2M3YXK0 A0A182MID6 A0A2M4AN04 A0A2M4BMD5 A0A2M4BMG8 A0A2M4CMM9 E0W075 A0A2M4CV75 A0A182JWI9 V4CJ86 A0A182U5L8 A0A182HIK6 Q0IFQ5 A0A182VIR1 A0A0L8GWX2 A0A2M4CWL9 A0A182TCB2 A0A1I8MBS7 V9KLF4 B4KF33 A0A084WFL1 T1P9T1 A0A182YQW7 B4LU17 A0A182GUK8 A0A1W4UIH1 Q7QER0 A0A2C9H898 A0A182QHV1 A0A1B6EMM7 A0A336KKH4 B4MWC8 A0A1Q3G445 H2VBJ8 U5EJF9 A0A023ETR5 A0A3P9LXR2 A0A3Q2EKU4 H3DJ31 A0A3P9KCV8 Q4RLY4 A0A3Q3VJP4 A0A2J7Q269 A0A3Q1ECR3 A0A3Q1GWD2 A0A1Y1KQA6 B5X2C0 A0A1B6KZM5 A0A3B5JZF5 A0A1S2ZMU0 A0A1I8Q2L8 A0A182PFC6 A0A3Q3ESC1 A0A2D0RRD1 A0A3Q1C556 A0A3Q0RFW6 B3N4W9 Q9VMQ5 A0A1B0FH47 D3TNK7 A0A1A9UQZ6
EC Number
1.14.13.-
Pubmed
26227816
28756777
26354079
22118469
23622113
24508170
+ More
20075255 28648823 20798317 30249741 21347285 18362917 19820115 20920257 23761445 17994087 24845553 20566863 23254933 17510324 25315136 24402279 24438588 25244985 26483478 12364791 14747013 17210077 21551351 24945155 17554307 15496914 28004739 20433749 10731132 12537572 12537569 20353571
20075255 28648823 20798317 30249741 21347285 18362917 19820115 20920257 23761445 17994087 24845553 20566863 23254933 17510324 25315136 24402279 24438588 25244985 26483478 12364791 14747013 17210077 21551351 24945155 17554307 15496914 28004739 20433749 10731132 12537572 12537569 20353571
EMBL
JTDY01006890
KOB65613.1
ODYU01003165
SOQ41566.1
KZ150014
PZC75019.1
+ More
NWSH01004460 PCG65079.1 KQ460045 KPJ18145.1 AGBW02008891 OWR52288.1 GAIX01005666 JAA86894.1 KQ459604 KPI92350.1 KK107185 EZA55835.1 AAZX01007490 NNAY01002421 OXU21307.1 JR047411 AEY60411.1 KZ288322 PBC28175.1 GL453666 EFN75738.1 KQ434809 KZC06392.1 QOIP01000012 RLU15929.1 KQ435719 KOX78758.1 ADTU01016098 KQ761039 OAD58343.1 KQ981636 KYN38799.1 KQ976725 KYM76558.1 KQ978501 KYM93567.1 GBYB01009844 JAG79611.1 KQ982813 KYQ50311.1 KQ414704 KOC63288.1 KQ978957 KYN27226.1 KQ971353 KYB26591.1 GL441504 EFN64713.1 ADMH02000338 ETN66912.1 CH916368 EDW03828.1 KK853628 KDR03814.1 GDRN01069811 JAI63990.1 GGFM01003794 MBW24545.1 GGFM01000242 MBW20993.1 AXCM01004049 GGFK01008854 MBW42175.1 GGFJ01005013 MBW54154.1 GGFJ01005012 MBW54153.1 GGFL01001970 MBW66148.1 AAZO01006645 DS235857 EEB19031.1 GGFL01005069 MBW69247.1 KB200294 ESP02270.1 APCN01003993 CH477294 EAT44481.1 KQ420065 KOF81417.1 GGFL01005070 MBW69248.1 JW866370 AFO98887.1 CH933807 EDW13016.1 ATLV01023372 KE525342 KFB49005.1 KA645511 AFP60140.1 CH940649 EDW65070.1 JXUM01089328 KQ563763 KXJ73351.1 AAAB01008846 EAA06342.5 AXCN02002023 GECZ01030618 JAS39151.1 UFQS01000499 UFQT01000499 SSX04418.1 SSX24782.1 CH963857 EDW75998.1 GFDL01000474 JAV34571.1 GANO01002249 JAB57622.1 GAPW01001789 JAC11809.1 CAAE01015019 CAG10598.1 NEVH01019373 PNF22683.1 GEZM01079905 GEZM01079904 JAV62350.1 BT045189 ACI33451.1 GEBQ01023075 JAT16902.1 CH954177 EDV57871.1 AE014134 AY058443 CCAG010021907 EZ423009 ADD19285.1
NWSH01004460 PCG65079.1 KQ460045 KPJ18145.1 AGBW02008891 OWR52288.1 GAIX01005666 JAA86894.1 KQ459604 KPI92350.1 KK107185 EZA55835.1 AAZX01007490 NNAY01002421 OXU21307.1 JR047411 AEY60411.1 KZ288322 PBC28175.1 GL453666 EFN75738.1 KQ434809 KZC06392.1 QOIP01000012 RLU15929.1 KQ435719 KOX78758.1 ADTU01016098 KQ761039 OAD58343.1 KQ981636 KYN38799.1 KQ976725 KYM76558.1 KQ978501 KYM93567.1 GBYB01009844 JAG79611.1 KQ982813 KYQ50311.1 KQ414704 KOC63288.1 KQ978957 KYN27226.1 KQ971353 KYB26591.1 GL441504 EFN64713.1 ADMH02000338 ETN66912.1 CH916368 EDW03828.1 KK853628 KDR03814.1 GDRN01069811 JAI63990.1 GGFM01003794 MBW24545.1 GGFM01000242 MBW20993.1 AXCM01004049 GGFK01008854 MBW42175.1 GGFJ01005013 MBW54154.1 GGFJ01005012 MBW54153.1 GGFL01001970 MBW66148.1 AAZO01006645 DS235857 EEB19031.1 GGFL01005069 MBW69247.1 KB200294 ESP02270.1 APCN01003993 CH477294 EAT44481.1 KQ420065 KOF81417.1 GGFL01005070 MBW69248.1 JW866370 AFO98887.1 CH933807 EDW13016.1 ATLV01023372 KE525342 KFB49005.1 KA645511 AFP60140.1 CH940649 EDW65070.1 JXUM01089328 KQ563763 KXJ73351.1 AAAB01008846 EAA06342.5 AXCN02002023 GECZ01030618 JAS39151.1 UFQS01000499 UFQT01000499 SSX04418.1 SSX24782.1 CH963857 EDW75998.1 GFDL01000474 JAV34571.1 GANO01002249 JAB57622.1 GAPW01001789 JAC11809.1 CAAE01015019 CAG10598.1 NEVH01019373 PNF22683.1 GEZM01079905 GEZM01079904 JAV62350.1 BT045189 ACI33451.1 GEBQ01023075 JAT16902.1 CH954177 EDV57871.1 AE014134 AY058443 CCAG010021907 EZ423009 ADD19285.1
Proteomes
UP000037510
UP000218220
UP000053240
UP000007151
UP000053268
UP000053097
+ More
UP000002358 UP000215335 UP000242457 UP000008237 UP000005203 UP000076502 UP000279307 UP000053105 UP000005205 UP000078541 UP000078540 UP000078542 UP000085678 UP000075809 UP000053825 UP000078492 UP000007266 UP000069272 UP000000311 UP000000673 UP000075884 UP000001070 UP000027135 UP000075900 UP000075883 UP000009046 UP000075881 UP000030746 UP000075902 UP000075840 UP000008820 UP000075903 UP000053454 UP000075901 UP000095301 UP000009192 UP000030765 UP000076408 UP000008792 UP000069940 UP000249989 UP000192221 UP000007062 UP000076407 UP000075886 UP000007798 UP000005226 UP000265180 UP000265020 UP000007303 UP000261620 UP000235965 UP000257200 UP000265040 UP000087266 UP000079721 UP000095300 UP000075885 UP000264800 UP000221080 UP000257160 UP000261340 UP000008711 UP000000803 UP000092444 UP000078200
UP000002358 UP000215335 UP000242457 UP000008237 UP000005203 UP000076502 UP000279307 UP000053105 UP000005205 UP000078541 UP000078540 UP000078542 UP000085678 UP000075809 UP000053825 UP000078492 UP000007266 UP000069272 UP000000311 UP000000673 UP000075884 UP000001070 UP000027135 UP000075900 UP000075883 UP000009046 UP000075881 UP000030746 UP000075902 UP000075840 UP000008820 UP000075903 UP000053454 UP000075901 UP000095301 UP000009192 UP000030765 UP000076408 UP000008792 UP000069940 UP000249989 UP000192221 UP000007062 UP000076407 UP000075886 UP000007798 UP000005226 UP000265180 UP000265020 UP000007303 UP000261620 UP000235965 UP000257200 UP000265040 UP000087266 UP000079721 UP000095300 UP000075885 UP000264800 UP000221080 UP000257160 UP000261340 UP000008711 UP000000803 UP000092444 UP000078200
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A0L7KQN6
A0A2H1VL88
A0A2W1BIY2
A0A2A4IZL9
A0A194RLF2
A0A212FEZ2
+ More
S4P958 A0A194PMK8 A0A026WIJ2 K7JED1 A0A232ESL8 V9IIU2 A0A2A3E8X6 E2C8K8 A0A088A361 A0A154P3J3 A0A3L8D6P3 A0A0N0U755 A0A158NI87 A0A310SI00 A0A195FFJ8 A0A195AWD4 A0A195BY98 A0A0C9R127 A0A1S3IS81 A0A151WR83 A0A0L7QXE3 A0A195EGE9 A0A139WFD9 A0A182FJL6 E2APB4 W5JRB1 A0A182N043 B4JAF7 A0A067QHI4 A0A182R6D5 A0A0P4WEC7 A0A2M3Z7R3 A0A2M3YXK0 A0A182MID6 A0A2M4AN04 A0A2M4BMD5 A0A2M4BMG8 A0A2M4CMM9 E0W075 A0A2M4CV75 A0A182JWI9 V4CJ86 A0A182U5L8 A0A182HIK6 Q0IFQ5 A0A182VIR1 A0A0L8GWX2 A0A2M4CWL9 A0A182TCB2 A0A1I8MBS7 V9KLF4 B4KF33 A0A084WFL1 T1P9T1 A0A182YQW7 B4LU17 A0A182GUK8 A0A1W4UIH1 Q7QER0 A0A2C9H898 A0A182QHV1 A0A1B6EMM7 A0A336KKH4 B4MWC8 A0A1Q3G445 H2VBJ8 U5EJF9 A0A023ETR5 A0A3P9LXR2 A0A3Q2EKU4 H3DJ31 A0A3P9KCV8 Q4RLY4 A0A3Q3VJP4 A0A2J7Q269 A0A3Q1ECR3 A0A3Q1GWD2 A0A1Y1KQA6 B5X2C0 A0A1B6KZM5 A0A3B5JZF5 A0A1S2ZMU0 A0A1I8Q2L8 A0A182PFC6 A0A3Q3ESC1 A0A2D0RRD1 A0A3Q1C556 A0A3Q0RFW6 B3N4W9 Q9VMQ5 A0A1B0FH47 D3TNK7 A0A1A9UQZ6
S4P958 A0A194PMK8 A0A026WIJ2 K7JED1 A0A232ESL8 V9IIU2 A0A2A3E8X6 E2C8K8 A0A088A361 A0A154P3J3 A0A3L8D6P3 A0A0N0U755 A0A158NI87 A0A310SI00 A0A195FFJ8 A0A195AWD4 A0A195BY98 A0A0C9R127 A0A1S3IS81 A0A151WR83 A0A0L7QXE3 A0A195EGE9 A0A139WFD9 A0A182FJL6 E2APB4 W5JRB1 A0A182N043 B4JAF7 A0A067QHI4 A0A182R6D5 A0A0P4WEC7 A0A2M3Z7R3 A0A2M3YXK0 A0A182MID6 A0A2M4AN04 A0A2M4BMD5 A0A2M4BMG8 A0A2M4CMM9 E0W075 A0A2M4CV75 A0A182JWI9 V4CJ86 A0A182U5L8 A0A182HIK6 Q0IFQ5 A0A182VIR1 A0A0L8GWX2 A0A2M4CWL9 A0A182TCB2 A0A1I8MBS7 V9KLF4 B4KF33 A0A084WFL1 T1P9T1 A0A182YQW7 B4LU17 A0A182GUK8 A0A1W4UIH1 Q7QER0 A0A2C9H898 A0A182QHV1 A0A1B6EMM7 A0A336KKH4 B4MWC8 A0A1Q3G445 H2VBJ8 U5EJF9 A0A023ETR5 A0A3P9LXR2 A0A3Q2EKU4 H3DJ31 A0A3P9KCV8 Q4RLY4 A0A3Q3VJP4 A0A2J7Q269 A0A3Q1ECR3 A0A3Q1GWD2 A0A1Y1KQA6 B5X2C0 A0A1B6KZM5 A0A3B5JZF5 A0A1S2ZMU0 A0A1I8Q2L8 A0A182PFC6 A0A3Q3ESC1 A0A2D0RRD1 A0A3Q1C556 A0A3Q0RFW6 B3N4W9 Q9VMQ5 A0A1B0FH47 D3TNK7 A0A1A9UQZ6
PDB
4N9X
E-value=1.07632e-08,
Score=138
Ontologies
GO
Topology
Subcellular location
Mitochondrion inner membrane
Golgi apparatus Localizes to cell processes and Golgi apparatus in podocytes. With evidence from 1 publications.
Cell projection Localizes to cell processes and Golgi apparatus in podocytes. With evidence from 1 publications.
Golgi apparatus Localizes to cell processes and Golgi apparatus in podocytes. With evidence from 1 publications.
Cell projection Localizes to cell processes and Golgi apparatus in podocytes. With evidence from 1 publications.
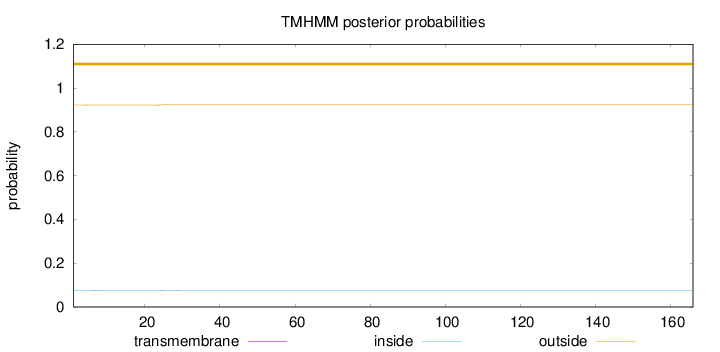
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05208
Exp number, first 60 AAs:
0.05208
Total prob of N-in:
0.07762
outside
1 - 166
Population Genetic Test Statistics
Pi
190.263006
Theta
167.157883
Tajima's D
0.44389
CLR
0
CSRT
0.502774861256937
Interpretation
Uncertain
