Gene
KWMTBOMO15225
Annotation
hemolymph_proteinase_5_[Danaus_plexippus]
Full name
Melanization protease 1
Location in the cell
Extracellular Reliability : 2.585
Sequence
CDS
ATGCTGCAGACGTTTTTTTTAGTCGTCACATCGATCCTGCAAATACAAAGTTTAGAATTTGGAGAGAGTTGTTTTGCAAATGATGAACCGGGAACGTGTATCGCCCTCTCAGAATGCAAGCCAGTTTTATCATACATCGAAAAGTTTGGAAATCAAATCCCTAATAATTTGCTACAGAAGTTTCAAGATCTGACGTGCGGCTATGACCACAGCGATCCCCTGGTTTGCTGCACTACATCCCAACACATAGCTGGATACTTTAACAATTTTGGTAATCAAAATGGCGGGACTAGCGTACCGAAACGACCGAAGCCCGTTCCTACTAGTAACAGCGGTAATTGGGGCAGTCAACCAAGCAAGGAACAAACAACAAGCAACCGACCGACTACTGCTGATACTGATATTAATAAGAGCGGCCGTGATGACGACTTTCCGGACATTCGTCAGAACCCTAACTTGAGTTTACTATCAAGTAATTGTGGAACGATAGAAAGCGATAGAATATTCGGAGGGAATCGTACCCGACTGTTCGAGATGCCGTGGATGGTGCTTCTTTCCTACGATTCAGATCGTGGAACAAAACTGAATTGCGGAGGAACACTTATAAACGAATGGTATGTTTTAACGGCCGCGCATTGTGTATCGTTCTTAGGAAATAGATTAACATTAAAACACGTTATCCTGGGAGAATATGACACAAGGCAAGATCCTGACTGCGAAAGATCGGAAGGAAAGGAATATTGCGCGGCTGGTATAATCACGGCAGAATTCGACGAAGTAATTCCTCATACAGGCTACACACCACAAACATTAATCGATGATATAGCATTAATCAGACTGAAAGAGCCTGCCGACTTTGATTTAGATAATATTAAAGCGATTTGCTTACCGATTACTCCTGAACTGCAAAGGGAGCCTCTTGTCGATAACTTTGGCGTGGTAGCAGGTTGGGGAGTGACAGAAGAAGGACTACAATCTCCTGTTTTGCTTAGTGTACAGCTGCCAATTCTCAGCAAAGAAAAATGTTTGCAAGACTATTCCCAATATTCCTTGAAAATCAACGACAAGCAACTCTGCGCCGGCGGTTTGCACGATAAGGATTCATGCGCCGGGGATTCTGGTGGCCCTTTGCTGTATCCAGGGAAAGTTGGCTCAACTGGCGTCCGATATGTTCAGAGAGGCATAGTCTCGTTTGGGTCGAAGCGGTGTGGTATCAGCCTGCCGGGAGTGTACACGAATGTTGCATACTACATGGATTGGATCTTGAATAATATTAGAAGTTGA
Protein
MLQTFFLVVTSILQIQSLEFGESCFANDEPGTCIALSECKPVLSYIEKFGNQIPNNLLQKFQDLTCGYDHSDPLVCCTTSQHIAGYFNNFGNQNGGTSVPKRPKPVPTSNSGNWGSQPSKEQTTSNRPTTADTDINKSGRDDDFPDIRQNPNLSLLSSNCGTIESDRIFGGNRTRLFEMPWMVLLSYDSDRGTKLNCGGTLINEWYVLTAAHCVSFLGNRLTLKHVILGEYDTRQDPDCERSEGKEYCAAGIITAEFDEVIPHTGYTPQTLIDDIALIRLKEPADFDLDNIKAICLPITPELQREPLVDNFGVVAGWGVTEEGLQSPVLLSVQLPILSKEKCLQDYSQYSLKINDKQLCAGGLHDKDSCAGDSGGPLLYPGKVGSTGVRYVQRGIVSFGSKRCGISLPGVYTNVAYYMDWILNNIRS
Summary
Description
Serine protease which plays an essential role in the melanization immune response by acting downstream of sp7 to activate prophenoloxidase (PPO1) (PubMed:16861233). May function in diverse Hayan-dependent PPO1-activating cascades that are negatively controlled by different serpin proteins; Spn27A in the hemolymph and Spn77BA in the trachea (PubMed:18854145, PubMed:16861233). Regulation of melanization and PPO1 activation appears to be largely independent of the Toll signaling pathway (PubMed:16861233).
Similarity
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family. CLIP subfamily.
Belongs to the peptidase S1 family. CLIP subfamily.
Keywords
Alternative splicing
Calcium
Complete proteome
Disulfide bond
Glycoprotein
Hydrolase
Metal-binding
Protease
Reference proteome
Serine protease
Signal
Zymogen
Feature
propeptide Activation peptide
chain Melanization protease 1
splice variant In isoform A.
chain Melanization protease 1
splice variant In isoform A.
Uniprot
A0A212F937
G9F9I4
A0A2H1W4A8
A0A2A4IZ28
A0A2W1BQ49
A0A194PG32
+ More
I4DNT5 A0A194RL52 Q5MPC9 A0A2J7Q286 A0A2P8YP04 A0A1L8DR27 A0A1L8DQP3 A0A1V1G586 A0A0M9A8Q5 A0A067QFH0 D6WP83 A0A0L7QXJ6 A0A026WIG3 A0A195FEJ5 F4WPR7 A0A195EG59 A0A0C9PIF6 E2C8L1 U5EMJ9 A0A0L7LRR0 D6WP86 A0A3L8D7J0 K7J981 E2C8L2 A0A232EZG2 A0A1B0CMU1 A0A1W6EWF7 A0A2J7Q299 A0A182QQA1 A0A154P3E5 A0A310SR39 A0A182S526 A0A1Y1MBK3 F4WPR8 A0A1V1FKN6 A0A139WF86 A0A0J7NWE6 A0A3F2YU18 E0VXK9 A0A182YKD4 A0A034VLF9 K7J980 Q7QKL1 A0A182MDC4 E2AV85 A0A195EG37 A0A2C9GV88 B4PNV6 A0A1I8JTD0 I4DKD5 A0A154P5I7 A0A182V2M2 A0A0M9A7E2 A0A0L7QXF0 A0A182XUW3 A0A1Y1L107 A0A126GUP6-2 A0A0C9QZC0 A0A034VVW3 U4U606 A0A067QHJ6 A0A0A1WS20 J3JYT9 B4QUZ8 A0A1W4UJW8 A0A1D2MDI9 A0A0C9QBV2
I4DNT5 A0A194RL52 Q5MPC9 A0A2J7Q286 A0A2P8YP04 A0A1L8DR27 A0A1L8DQP3 A0A1V1G586 A0A0M9A8Q5 A0A067QFH0 D6WP83 A0A0L7QXJ6 A0A026WIG3 A0A195FEJ5 F4WPR7 A0A195EG59 A0A0C9PIF6 E2C8L1 U5EMJ9 A0A0L7LRR0 D6WP86 A0A3L8D7J0 K7J981 E2C8L2 A0A232EZG2 A0A1B0CMU1 A0A1W6EWF7 A0A2J7Q299 A0A182QQA1 A0A154P3E5 A0A310SR39 A0A182S526 A0A1Y1MBK3 F4WPR8 A0A1V1FKN6 A0A139WF86 A0A0J7NWE6 A0A3F2YU18 E0VXK9 A0A182YKD4 A0A034VLF9 K7J980 Q7QKL1 A0A182MDC4 E2AV85 A0A195EG37 A0A2C9GV88 B4PNV6 A0A1I8JTD0 I4DKD5 A0A154P5I7 A0A182V2M2 A0A0M9A7E2 A0A0L7QXF0 A0A182XUW3 A0A1Y1L107 A0A126GUP6-2 A0A0C9QZC0 A0A034VVW3 U4U606 A0A067QHJ6 A0A0A1WS20 J3JYT9 B4QUZ8 A0A1W4UJW8 A0A1D2MDI9 A0A0C9QBV2
EC Number
3.4.21.-
Pubmed
22118469
28756777
26354079
22651552
15944088
29403074
+ More
28410430 24845553 18362917 19820115 24508170 21719571 20798317 26227816 30249741 20075255 28648823 28004739 20566863 25244985 25348373 12364791 14747013 17210077 17994087 17550304 10731132 12537572 10731138 16861233 18854145 22227521 23537049 25830018 22516182 27289101
28410430 24845553 18362917 19820115 24508170 21719571 20798317 26227816 30249741 20075255 28648823 28004739 20566863 25244985 25348373 12364791 14747013 17210077 17994087 17550304 10731132 12537572 10731138 16861233 18854145 22227521 23537049 25830018 22516182 27289101
EMBL
AGBW02009640
OWR50265.1
JN033729
AEW46882.1
ODYU01006253
SOQ47925.1
+ More
NWSH01004460 PCG65075.1 KZ150014 PZC75016.1 KQ459604 KPI92346.1 AK403129 BAM19575.1 KQ460045 KPJ18149.1 AY672781 AAV91003.1 NEVH01019373 PNF22687.1 PYGN01000457 PSN45975.1 GFDF01005299 JAV08785.1 GFDF01005298 JAV08786.1 FX985343 BAX07356.1 KQ435719 KOX78756.1 KK853628 KDR03823.1 KQ971353 EFA07454.1 KQ414704 KOC63286.1 KK107185 EZA55837.1 KQ981636 KYN38801.1 GL888255 EGI63808.1 KQ978957 KYN27228.1 GBYB01000710 GBYB01000713 JAG70477.1 JAG70480.1 GL453666 EFN75741.1 GANO01000994 JAB58877.1 JTDY01000242 KOB78094.1 EFA07452.2 QOIP01000012 RLU16063.1 AAZX01002213 EFN75742.1 NNAY01001477 OXU23832.1 AJWK01019165 AJWK01019166 AJWK01019167 AJWK01019168 KY563640 ARK20049.1 PNF22708.1 AXCN02000165 KQ434809 KZC06391.1 KQ761039 OAD58341.1 GEZM01037774 JAV81940.1 EGI63809.1 FX985369 BAX07382.1 KYB26592.1 LBMM01001207 KMQ96720.1 DS235833 EEB18115.1 GAKP01015995 JAC42957.1 AAAB01008794 EAA03537.4 AXCM01000867 GL443059 EFN62639.1 KYN27230.1 CM000160 EDW98166.2 APCN01002147 AK401753 BAM18375.1 KZC06390.1 KOX78755.1 KOC63285.1 GEZM01073843 JAV64787.1 AE014297 AF145611 BT125977 AAD38586.1 ADX35956.1 GBYB01001065 JAG70832.1 GAKP01013295 GAKP01013294 JAC45658.1 KB631684 ERL85365.1 KDR03824.1 GBXI01012856 JAD01436.1 BT128420 AEE63377.1 CM000364 EDX11579.1 LJIJ01001731 ODM90951.1 GBYB01000714 GBYB01006695 JAG70481.1 JAG76462.1
NWSH01004460 PCG65075.1 KZ150014 PZC75016.1 KQ459604 KPI92346.1 AK403129 BAM19575.1 KQ460045 KPJ18149.1 AY672781 AAV91003.1 NEVH01019373 PNF22687.1 PYGN01000457 PSN45975.1 GFDF01005299 JAV08785.1 GFDF01005298 JAV08786.1 FX985343 BAX07356.1 KQ435719 KOX78756.1 KK853628 KDR03823.1 KQ971353 EFA07454.1 KQ414704 KOC63286.1 KK107185 EZA55837.1 KQ981636 KYN38801.1 GL888255 EGI63808.1 KQ978957 KYN27228.1 GBYB01000710 GBYB01000713 JAG70477.1 JAG70480.1 GL453666 EFN75741.1 GANO01000994 JAB58877.1 JTDY01000242 KOB78094.1 EFA07452.2 QOIP01000012 RLU16063.1 AAZX01002213 EFN75742.1 NNAY01001477 OXU23832.1 AJWK01019165 AJWK01019166 AJWK01019167 AJWK01019168 KY563640 ARK20049.1 PNF22708.1 AXCN02000165 KQ434809 KZC06391.1 KQ761039 OAD58341.1 GEZM01037774 JAV81940.1 EGI63809.1 FX985369 BAX07382.1 KYB26592.1 LBMM01001207 KMQ96720.1 DS235833 EEB18115.1 GAKP01015995 JAC42957.1 AAAB01008794 EAA03537.4 AXCM01000867 GL443059 EFN62639.1 KYN27230.1 CM000160 EDW98166.2 APCN01002147 AK401753 BAM18375.1 KZC06390.1 KOX78755.1 KOC63285.1 GEZM01073843 JAV64787.1 AE014297 AF145611 BT125977 AAD38586.1 ADX35956.1 GBYB01001065 JAG70832.1 GAKP01013295 GAKP01013294 JAC45658.1 KB631684 ERL85365.1 KDR03824.1 GBXI01012856 JAD01436.1 BT128420 AEE63377.1 CM000364 EDX11579.1 LJIJ01001731 ODM90951.1 GBYB01000714 GBYB01006695 JAG70481.1 JAG76462.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000235965
UP000245037
+ More
UP000053105 UP000027135 UP000007266 UP000053825 UP000053097 UP000078541 UP000007755 UP000078492 UP000008237 UP000037510 UP000279307 UP000002358 UP000215335 UP000092461 UP000075886 UP000076502 UP000075900 UP000036403 UP000075881 UP000009046 UP000076408 UP000007062 UP000075883 UP000000311 UP000075884 UP000002282 UP000075840 UP000075903 UP000076407 UP000000803 UP000030742 UP000000304 UP000192221 UP000094527
UP000053105 UP000027135 UP000007266 UP000053825 UP000053097 UP000078541 UP000007755 UP000078492 UP000008237 UP000037510 UP000279307 UP000002358 UP000215335 UP000092461 UP000075886 UP000076502 UP000075900 UP000036403 UP000075881 UP000009046 UP000076408 UP000007062 UP000075883 UP000000311 UP000075884 UP000002282 UP000075840 UP000075903 UP000076407 UP000000803 UP000030742 UP000000304 UP000192221 UP000094527
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A212F937
G9F9I4
A0A2H1W4A8
A0A2A4IZ28
A0A2W1BQ49
A0A194PG32
+ More
I4DNT5 A0A194RL52 Q5MPC9 A0A2J7Q286 A0A2P8YP04 A0A1L8DR27 A0A1L8DQP3 A0A1V1G586 A0A0M9A8Q5 A0A067QFH0 D6WP83 A0A0L7QXJ6 A0A026WIG3 A0A195FEJ5 F4WPR7 A0A195EG59 A0A0C9PIF6 E2C8L1 U5EMJ9 A0A0L7LRR0 D6WP86 A0A3L8D7J0 K7J981 E2C8L2 A0A232EZG2 A0A1B0CMU1 A0A1W6EWF7 A0A2J7Q299 A0A182QQA1 A0A154P3E5 A0A310SR39 A0A182S526 A0A1Y1MBK3 F4WPR8 A0A1V1FKN6 A0A139WF86 A0A0J7NWE6 A0A3F2YU18 E0VXK9 A0A182YKD4 A0A034VLF9 K7J980 Q7QKL1 A0A182MDC4 E2AV85 A0A195EG37 A0A2C9GV88 B4PNV6 A0A1I8JTD0 I4DKD5 A0A154P5I7 A0A182V2M2 A0A0M9A7E2 A0A0L7QXF0 A0A182XUW3 A0A1Y1L107 A0A126GUP6-2 A0A0C9QZC0 A0A034VVW3 U4U606 A0A067QHJ6 A0A0A1WS20 J3JYT9 B4QUZ8 A0A1W4UJW8 A0A1D2MDI9 A0A0C9QBV2
I4DNT5 A0A194RL52 Q5MPC9 A0A2J7Q286 A0A2P8YP04 A0A1L8DR27 A0A1L8DQP3 A0A1V1G586 A0A0M9A8Q5 A0A067QFH0 D6WP83 A0A0L7QXJ6 A0A026WIG3 A0A195FEJ5 F4WPR7 A0A195EG59 A0A0C9PIF6 E2C8L1 U5EMJ9 A0A0L7LRR0 D6WP86 A0A3L8D7J0 K7J981 E2C8L2 A0A232EZG2 A0A1B0CMU1 A0A1W6EWF7 A0A2J7Q299 A0A182QQA1 A0A154P3E5 A0A310SR39 A0A182S526 A0A1Y1MBK3 F4WPR8 A0A1V1FKN6 A0A139WF86 A0A0J7NWE6 A0A3F2YU18 E0VXK9 A0A182YKD4 A0A034VLF9 K7J980 Q7QKL1 A0A182MDC4 E2AV85 A0A195EG37 A0A2C9GV88 B4PNV6 A0A1I8JTD0 I4DKD5 A0A154P5I7 A0A182V2M2 A0A0M9A7E2 A0A0L7QXF0 A0A182XUW3 A0A1Y1L107 A0A126GUP6-2 A0A0C9QZC0 A0A034VVW3 U4U606 A0A067QHJ6 A0A0A1WS20 J3JYT9 B4QUZ8 A0A1W4UJW8 A0A1D2MDI9 A0A0C9QBV2
PDB
2XXL
E-value=2.19343e-46,
Score=468
Ontologies
GO
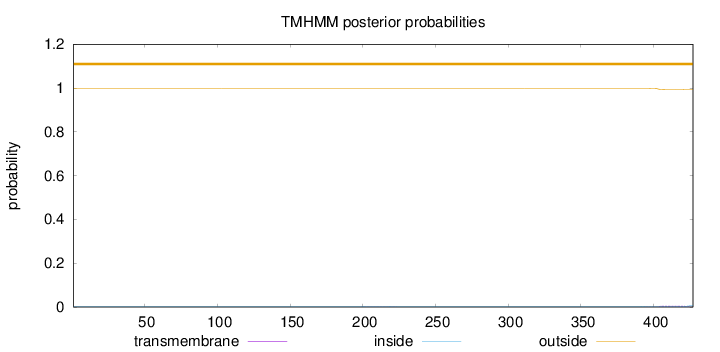
Topology
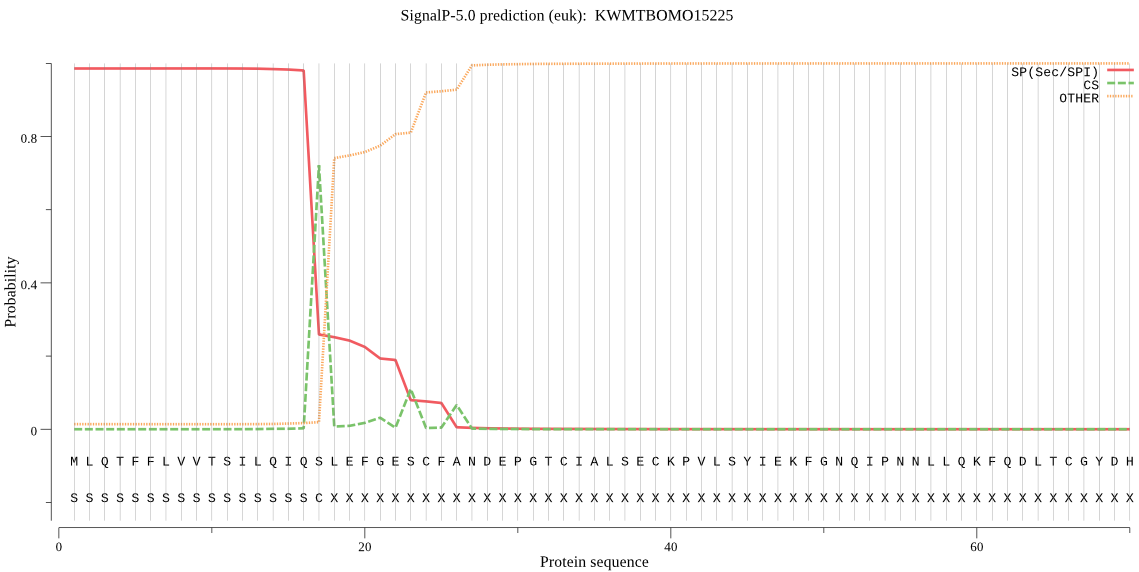
SignalP
Position: 1 - 17,
Likelihood: 0.986002
Length:
427
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13038
Exp number, first 60 AAs:
0.00183
Total prob of N-in:
0.00191
outside
1 - 427
Population Genetic Test Statistics
Pi
189.874502
Theta
178.233668
Tajima's D
1.511313
CLR
0
CSRT
0.789660516974151
Interpretation
Uncertain
