Gene
KWMTBOMO15223
Pre Gene Modal
BGIBMGA005006
Annotation
PREDICTED:_uncharacterized_protein_C14orf119_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.577 Extracellular Reliability : 1.094
Sequence
CDS
ATGTCTGCGCAAGGCCTAACCTCTGAAGCTCAATTGCGATACCTTTTACAATGGTTTGGAGAGTTTAGTGAACTACAGCGTGAAGATTTTTTGCCGGTACTTGCTGCATCACGATCTGATAACCCTGAACAATTGGCCGCTACCATGGCAAATATGACTTGTCAAGATAAACCGGTCAGCCTATTTCAGTGTCGAATCAAACTATTCAATGAATGGTTTCCAACTTGGTCTCGGGAAGAACAGGACAGATTTGTCAATAAAATATCTGAAATAGACTCCGAATTTGGTGAGAAACTTCACGAAATAATCACTGATGGGCCTAAAGTTAATGGAACTGATGAACCTTGGATCAATGAGCAGAATGTAGAACCCGAACGCGAGGATCAGGTACCAGAAAACATTCCTGTTGAAATTGTAGCTGCTTCATAA
Protein
MSAQGLTSEAQLRYLLQWFGEFSELQREDFLPVLAASRSDNPEQLAATMANMTCQDKPVSLFQCRIKLFNEWFPTWSREEQDRFVNKISEIDSEFGEKLHEIITDGPKVNGTDEPWINEQNVEPEREDQVPENIPVEIVAAS
Summary
Similarity
Belongs to the Glu/Leu/Phe/Val dehydrogenases family.
Uniprot
H9J665
A0A2H1VEC7
A0A2A4IW78
A0A2A4IXS5
A0A3S2NY35
A0A194PHZ3
+ More
A0A2W1BTF7 I4DN86 S4PWC3 A0A1E1WB10 A0A1E1WU17 A0A212EQZ5 A0A194RKT6 A0A0T6BB97 D6WSS4 A0A023EH09 A0A023EI71 A0A1B0DE21 N6TPM7 A0A182G582 Q17B78 A0A1S4F9H9 A0A1B0CSV3 W8CDF3 A0A1Q3F307 A0A1Q3F2Z2 A0A1W4W0C4 A0A1Q3F3W3 A0A2S2P7G1 Q6NL99 A0A2H8TD41 A0A182T343 B0WQE5 A0A182YGL6 B3NZL1 B4HJU1 Q9VH82 B4QVZ3 A0A182LYB8 A0A182RYL7 J9JSG1 A0A2M4CY54 A0A0A1WHK1 A0A084WTM4 A0A2M3ZDF7 A0A0K8WDL5 A0A034WC36 A0A2M4AY67 A0A2S2R038 B4PVE1 A0A026WZB2 Q299G7 B4G555 B4N7Z4 A0A2M4CYA9 A0A182N5R6 A0A1I8PYK1 A0A182W1J8 B3MTY4 W5J8S2 A0A2R7W827 A0A0J7NEN0 A0A182IQG9 A0A182PPY5 A0A0L7KMP9 A0A2M4CUS3 A0A1B6GMN2 A0A1B6JVY4 A0A2P8YI04 A0A182FPJ0 A0A1I8NG82 T1PI46 Q5TTE3 A0A3B0K504 A0A3S4R6U3 A0A2M4C188 A0A2M4C0C1 A0A2M4C0A3 A0A1B6KG89 A0A182I7U2 A0A182XJ14 A0A182QRE1 A0A182JPR2 A0A182UKW7 A0A224YDR7 A0A182L7E1 E9IUN3 A0A151X020 A0A195DVK1 A0A195EXM7 A0A067RK48 T1JLH5 B4JV73 A0A1A9YQ08 A0A0L0CM60 D3TQB1 K7J956 A0A1A9WC16 A0A0M4EM19
A0A2W1BTF7 I4DN86 S4PWC3 A0A1E1WB10 A0A1E1WU17 A0A212EQZ5 A0A194RKT6 A0A0T6BB97 D6WSS4 A0A023EH09 A0A023EI71 A0A1B0DE21 N6TPM7 A0A182G582 Q17B78 A0A1S4F9H9 A0A1B0CSV3 W8CDF3 A0A1Q3F307 A0A1Q3F2Z2 A0A1W4W0C4 A0A1Q3F3W3 A0A2S2P7G1 Q6NL99 A0A2H8TD41 A0A182T343 B0WQE5 A0A182YGL6 B3NZL1 B4HJU1 Q9VH82 B4QVZ3 A0A182LYB8 A0A182RYL7 J9JSG1 A0A2M4CY54 A0A0A1WHK1 A0A084WTM4 A0A2M3ZDF7 A0A0K8WDL5 A0A034WC36 A0A2M4AY67 A0A2S2R038 B4PVE1 A0A026WZB2 Q299G7 B4G555 B4N7Z4 A0A2M4CYA9 A0A182N5R6 A0A1I8PYK1 A0A182W1J8 B3MTY4 W5J8S2 A0A2R7W827 A0A0J7NEN0 A0A182IQG9 A0A182PPY5 A0A0L7KMP9 A0A2M4CUS3 A0A1B6GMN2 A0A1B6JVY4 A0A2P8YI04 A0A182FPJ0 A0A1I8NG82 T1PI46 Q5TTE3 A0A3B0K504 A0A3S4R6U3 A0A2M4C188 A0A2M4C0C1 A0A2M4C0A3 A0A1B6KG89 A0A182I7U2 A0A182XJ14 A0A182QRE1 A0A182JPR2 A0A182UKW7 A0A224YDR7 A0A182L7E1 E9IUN3 A0A151X020 A0A195DVK1 A0A195EXM7 A0A067RK48 T1JLH5 B4JV73 A0A1A9YQ08 A0A0L0CM60 D3TQB1 K7J956 A0A1A9WC16 A0A0M4EM19
Pubmed
19121390
26354079
28756777
22651552
23622113
22118469
+ More
18362917 19820115 24945155 23537049 26483478 17510324 24495485 25244985 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24438588 25348373 17550304 24508170 30249741 15632085 20920257 23761445 26227816 29403074 25315136 12364791 14747013 17210077 28797301 20966253 21282665 24845553 26108605 20353571 20075255
18362917 19820115 24945155 23537049 26483478 17510324 24495485 25244985 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24438588 25348373 17550304 24508170 30249741 15632085 20920257 23761445 26227816 29403074 25315136 12364791 14747013 17210077 28797301 20966253 21282665 24845553 26108605 20353571 20075255
EMBL
BABH01019991
ODYU01001850
SOQ38624.1
NWSH01005677
PCG64065.1
PCG64064.1
+ More
RSAL01000102 RVE47483.1 KQ459604 KPI92344.1 KZ150014 PZC75013.1 AK402772 BAM19376.1 GAIX01007038 JAA85522.1 GDQN01006864 JAT84190.1 GDQN01000718 JAT90336.1 AGBW02013167 OWR43918.1 KQ460045 KPJ18152.1 LJIG01002336 KRT84604.1 KQ971352 EFA06361.1 GAPW01004955 JAC08643.1 GAPW01004988 JAC08610.1 AJVK01032467 APGK01059368 KB741293 KB631893 ENN70231.1 ERL86921.1 JXUM01042960 JXUM01042961 JXUM01042962 KQ561344 KXJ78895.1 CH477325 EAT43489.1 AJWK01026648 AJWK01026649 GAMC01002046 JAC04510.1 GFDL01013112 JAV21933.1 GFDL01013119 JAV21926.1 GFDL01012836 JAV22209.1 GGMR01012741 MBY25360.1 BT012437 AAS93708.1 GFXV01000212 MBW12017.1 DS232039 EDS32816.1 CH954181 EDV49721.1 CH480815 EDW42825.1 AE014297 BT031188 AAF54437.1 ABY20429.1 CM000364 EDX13573.1 AXCM01005702 ABLF02038581 GGFL01006059 MBW70237.1 GBXI01016292 GBXI01007580 JAC97999.1 JAD06712.1 ATLV01026902 KE525420 KFB53568.1 GGFM01005780 MBW26531.1 GDHF01003160 GDHF01001068 JAI49154.1 JAI51246.1 GAKP01007061 GAKP01007060 JAC51892.1 GGFK01012412 MBW45733.1 GGMS01013539 MBY82742.1 CM000160 EDW96714.1 KK107078 QOIP01000011 EZA60484.1 RLU16447.1 CM000070 EAL27736.3 CH479179 EDW24721.1 CH964232 EDW81245.1 GGFL01006081 MBW70259.1 CH902623 EDV30265.1 ADMH02002100 ETN59199.1 KK854443 PTY15896.1 LBMM01006055 KMQ90975.1 JTDY01008853 KOB64361.1 GGFL01004733 MBW68911.1 GECZ01006071 JAS63698.1 GECU01036896 GECU01024209 GECU01021850 GECU01017760 GECU01004335 JAS70810.1 JAS83497.1 JAS85856.1 JAS89946.1 JAT03372.1 PYGN01000580 PSN43873.1 KA647810 AFP62439.1 AAAB01008880 EAL40654.4 OUUW01000013 SPP87782.1 NCKU01001361 NCKU01001360 RWS12310.1 RWS12319.1 GGFJ01009893 MBW59034.1 GGFJ01009601 MBW58742.1 GGFJ01009606 MBW58747.1 GEBQ01029531 JAT10446.1 APCN01002556 AXCN02001158 GFPF01004702 MAA15848.1 GL765999 EFZ15724.1 KQ982624 KYQ53697.1 KQ980322 KYN16589.1 KQ981948 KYN32634.1 KK852425 KDR24162.1 JH431312 CH916374 EDV91393.1 JRES01000196 KNC33365.1 EZ423613 ADD19889.1 AAZX01002219 CP012526 ALC45464.1
RSAL01000102 RVE47483.1 KQ459604 KPI92344.1 KZ150014 PZC75013.1 AK402772 BAM19376.1 GAIX01007038 JAA85522.1 GDQN01006864 JAT84190.1 GDQN01000718 JAT90336.1 AGBW02013167 OWR43918.1 KQ460045 KPJ18152.1 LJIG01002336 KRT84604.1 KQ971352 EFA06361.1 GAPW01004955 JAC08643.1 GAPW01004988 JAC08610.1 AJVK01032467 APGK01059368 KB741293 KB631893 ENN70231.1 ERL86921.1 JXUM01042960 JXUM01042961 JXUM01042962 KQ561344 KXJ78895.1 CH477325 EAT43489.1 AJWK01026648 AJWK01026649 GAMC01002046 JAC04510.1 GFDL01013112 JAV21933.1 GFDL01013119 JAV21926.1 GFDL01012836 JAV22209.1 GGMR01012741 MBY25360.1 BT012437 AAS93708.1 GFXV01000212 MBW12017.1 DS232039 EDS32816.1 CH954181 EDV49721.1 CH480815 EDW42825.1 AE014297 BT031188 AAF54437.1 ABY20429.1 CM000364 EDX13573.1 AXCM01005702 ABLF02038581 GGFL01006059 MBW70237.1 GBXI01016292 GBXI01007580 JAC97999.1 JAD06712.1 ATLV01026902 KE525420 KFB53568.1 GGFM01005780 MBW26531.1 GDHF01003160 GDHF01001068 JAI49154.1 JAI51246.1 GAKP01007061 GAKP01007060 JAC51892.1 GGFK01012412 MBW45733.1 GGMS01013539 MBY82742.1 CM000160 EDW96714.1 KK107078 QOIP01000011 EZA60484.1 RLU16447.1 CM000070 EAL27736.3 CH479179 EDW24721.1 CH964232 EDW81245.1 GGFL01006081 MBW70259.1 CH902623 EDV30265.1 ADMH02002100 ETN59199.1 KK854443 PTY15896.1 LBMM01006055 KMQ90975.1 JTDY01008853 KOB64361.1 GGFL01004733 MBW68911.1 GECZ01006071 JAS63698.1 GECU01036896 GECU01024209 GECU01021850 GECU01017760 GECU01004335 JAS70810.1 JAS83497.1 JAS85856.1 JAS89946.1 JAT03372.1 PYGN01000580 PSN43873.1 KA647810 AFP62439.1 AAAB01008880 EAL40654.4 OUUW01000013 SPP87782.1 NCKU01001361 NCKU01001360 RWS12310.1 RWS12319.1 GGFJ01009893 MBW59034.1 GGFJ01009601 MBW58742.1 GGFJ01009606 MBW58747.1 GEBQ01029531 JAT10446.1 APCN01002556 AXCN02001158 GFPF01004702 MAA15848.1 GL765999 EFZ15724.1 KQ982624 KYQ53697.1 KQ980322 KYN16589.1 KQ981948 KYN32634.1 KK852425 KDR24162.1 JH431312 CH916374 EDV91393.1 JRES01000196 KNC33365.1 EZ423613 ADD19889.1 AAZX01002219 CP012526 ALC45464.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
+ More
UP000007266 UP000092462 UP000019118 UP000030742 UP000069940 UP000249989 UP000008820 UP000092461 UP000192221 UP000075901 UP000002320 UP000076408 UP000008711 UP000001292 UP000000803 UP000000304 UP000075883 UP000075900 UP000007819 UP000030765 UP000002282 UP000053097 UP000279307 UP000001819 UP000008744 UP000007798 UP000075884 UP000095300 UP000075920 UP000007801 UP000000673 UP000036403 UP000075880 UP000075885 UP000037510 UP000245037 UP000069272 UP000095301 UP000007062 UP000268350 UP000285301 UP000075840 UP000076407 UP000075886 UP000075881 UP000075902 UP000075882 UP000075809 UP000078492 UP000078541 UP000027135 UP000001070 UP000092443 UP000037069 UP000002358 UP000091820 UP000092553
UP000007266 UP000092462 UP000019118 UP000030742 UP000069940 UP000249989 UP000008820 UP000092461 UP000192221 UP000075901 UP000002320 UP000076408 UP000008711 UP000001292 UP000000803 UP000000304 UP000075883 UP000075900 UP000007819 UP000030765 UP000002282 UP000053097 UP000279307 UP000001819 UP000008744 UP000007798 UP000075884 UP000095300 UP000075920 UP000007801 UP000000673 UP000036403 UP000075880 UP000075885 UP000037510 UP000245037 UP000069272 UP000095301 UP000007062 UP000268350 UP000285301 UP000075840 UP000076407 UP000075886 UP000075881 UP000075902 UP000075882 UP000075809 UP000078492 UP000078541 UP000027135 UP000001070 UP000092443 UP000037069 UP000002358 UP000091820 UP000092553
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9J665
A0A2H1VEC7
A0A2A4IW78
A0A2A4IXS5
A0A3S2NY35
A0A194PHZ3
+ More
A0A2W1BTF7 I4DN86 S4PWC3 A0A1E1WB10 A0A1E1WU17 A0A212EQZ5 A0A194RKT6 A0A0T6BB97 D6WSS4 A0A023EH09 A0A023EI71 A0A1B0DE21 N6TPM7 A0A182G582 Q17B78 A0A1S4F9H9 A0A1B0CSV3 W8CDF3 A0A1Q3F307 A0A1Q3F2Z2 A0A1W4W0C4 A0A1Q3F3W3 A0A2S2P7G1 Q6NL99 A0A2H8TD41 A0A182T343 B0WQE5 A0A182YGL6 B3NZL1 B4HJU1 Q9VH82 B4QVZ3 A0A182LYB8 A0A182RYL7 J9JSG1 A0A2M4CY54 A0A0A1WHK1 A0A084WTM4 A0A2M3ZDF7 A0A0K8WDL5 A0A034WC36 A0A2M4AY67 A0A2S2R038 B4PVE1 A0A026WZB2 Q299G7 B4G555 B4N7Z4 A0A2M4CYA9 A0A182N5R6 A0A1I8PYK1 A0A182W1J8 B3MTY4 W5J8S2 A0A2R7W827 A0A0J7NEN0 A0A182IQG9 A0A182PPY5 A0A0L7KMP9 A0A2M4CUS3 A0A1B6GMN2 A0A1B6JVY4 A0A2P8YI04 A0A182FPJ0 A0A1I8NG82 T1PI46 Q5TTE3 A0A3B0K504 A0A3S4R6U3 A0A2M4C188 A0A2M4C0C1 A0A2M4C0A3 A0A1B6KG89 A0A182I7U2 A0A182XJ14 A0A182QRE1 A0A182JPR2 A0A182UKW7 A0A224YDR7 A0A182L7E1 E9IUN3 A0A151X020 A0A195DVK1 A0A195EXM7 A0A067RK48 T1JLH5 B4JV73 A0A1A9YQ08 A0A0L0CM60 D3TQB1 K7J956 A0A1A9WC16 A0A0M4EM19
A0A2W1BTF7 I4DN86 S4PWC3 A0A1E1WB10 A0A1E1WU17 A0A212EQZ5 A0A194RKT6 A0A0T6BB97 D6WSS4 A0A023EH09 A0A023EI71 A0A1B0DE21 N6TPM7 A0A182G582 Q17B78 A0A1S4F9H9 A0A1B0CSV3 W8CDF3 A0A1Q3F307 A0A1Q3F2Z2 A0A1W4W0C4 A0A1Q3F3W3 A0A2S2P7G1 Q6NL99 A0A2H8TD41 A0A182T343 B0WQE5 A0A182YGL6 B3NZL1 B4HJU1 Q9VH82 B4QVZ3 A0A182LYB8 A0A182RYL7 J9JSG1 A0A2M4CY54 A0A0A1WHK1 A0A084WTM4 A0A2M3ZDF7 A0A0K8WDL5 A0A034WC36 A0A2M4AY67 A0A2S2R038 B4PVE1 A0A026WZB2 Q299G7 B4G555 B4N7Z4 A0A2M4CYA9 A0A182N5R6 A0A1I8PYK1 A0A182W1J8 B3MTY4 W5J8S2 A0A2R7W827 A0A0J7NEN0 A0A182IQG9 A0A182PPY5 A0A0L7KMP9 A0A2M4CUS3 A0A1B6GMN2 A0A1B6JVY4 A0A2P8YI04 A0A182FPJ0 A0A1I8NG82 T1PI46 Q5TTE3 A0A3B0K504 A0A3S4R6U3 A0A2M4C188 A0A2M4C0C1 A0A2M4C0A3 A0A1B6KG89 A0A182I7U2 A0A182XJ14 A0A182QRE1 A0A182JPR2 A0A182UKW7 A0A224YDR7 A0A182L7E1 E9IUN3 A0A151X020 A0A195DVK1 A0A195EXM7 A0A067RK48 T1JLH5 B4JV73 A0A1A9YQ08 A0A0L0CM60 D3TQB1 K7J956 A0A1A9WC16 A0A0M4EM19
Ontologies
PANTHER
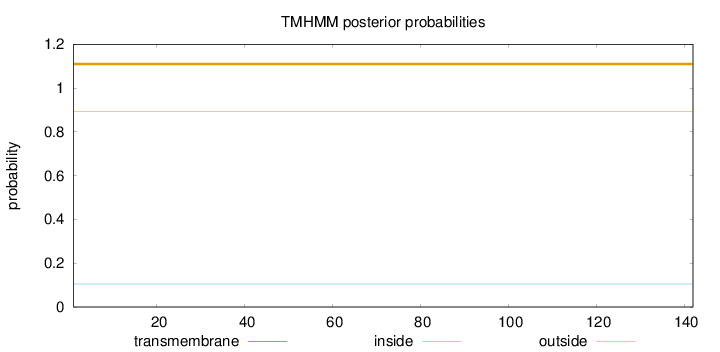
Topology
Length:
142
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10554
outside
1 - 142
Population Genetic Test Statistics
Pi
270.084473
Theta
203.948108
Tajima's D
0.629558
CLR
0
CSRT
0.545172741362932
Interpretation
Uncertain
