Pre Gene Modal
BGIBMGA005176
Annotation
PREDICTED:_synaptosomal-associated_protein_25_isoform_X4_[Papilio_polytes]
Full name
Synaptosomal-associated protein
+ More
Synaptosomal-associated protein 25
Synaptosomal-associated protein 25
Alternative Name
Synaptosomal-associated 25 kDa protein
Location in the cell
Nuclear Reliability : 2.904
Sequence
CDS
ATGCCTGCAGCAGTACCTCCTGCCGAAAATGGCGGGCCTCGCAGCGAGCTCGAGCAGCTGCAAATGCGAGCCGGTCAGGTTACCGATGAGTCGTTGGAGTCCACAAGGCGTATGATGCAACTATGTGAGGAGAGCAAGGAGGCCGGCATTCGTACTCTGGTCGCTCTAGACGACCAGGGAGAACAATTGGACAGAATCGAGGAGGGCATGGACCAGATCAACGCGGACATGCGGGAGGCCGAGAAGAATCTGTCCGGGATGGAGAAATGTTGCGGGATCTGCGTGCTGCCTTGCAACAAGGGAGCATCGTTCAAAGAGGACGACGGCACCTGGAAGGGCAACGATGACGGTAAGGTGGTCAACAATCAGCCGCAGCGGGTGATGGATGAACGCAATGGGATCGGCCCGCAGGCTGGATATATCGGGAGGATAACGAACGACGCTCGTGAAGACGAAATGGAGGAGAACATGGGTCAAGTGAACACCATGATCGGGAATCTGCGCAACATGGCCTTGGACATGGGTTCCGAGCTCGAGAACCAGAACCGACAGATCGACCGCATCAATCGCAAGGGAGAGTCAAATGAAACGAGGATAACTCTGGCGAACCAGCGCGCTCACGAGCTCCTCAAGTAA
Protein
MPAAVPPAENGGPRSELEQLQMRAGQVTDESLESTRRMMQLCEESKEAGIRTLVALDDQGEQLDRIEEGMDQINADMREAEKNLSGMEKCCGICVLPCNKGASFKEDDGTWKGNDDGKVVNNQPQRVMDERNGIGPQAGYIGRITNDAREDEMEENMGQVNTMIGNLRNMALDMGSELENQNRQIDRINRKGESNETRITLANQRAHELLK
Summary
Description
May play an important role in the synaptic function of specific neuronal systems. Associates with proteins involved in vesicle docking and membrane fusion (By similarity).
Similarity
Belongs to the SNAP-25 family.
Keywords
Alternative splicing
Cell junction
Coiled coil
Complete proteome
Reference proteome
Repeat
Synapse
Synaptosome
Feature
chain Synaptosomal-associated protein 25
splice variant In isoform C.
splice variant In isoform C.
Uniprot
A0A212F742
A0A194PHY8
A0A2W1BL53
A0A2A4K3F5
A0A182X9T1
F5HMS4
+ More
A0A1Y9IXP0 A0A1L8DVY8 A0A158NHW8 A0A1W4WS64 A0A1Y9IXY3 Q7PUS1 U5EIP5 E0VV38 A0A1Y9IY75 A0A1W7R879 A0A182RNH4 W8CEE5 A0A1W4WRW1 A0A1B6ENN8 A0A1I8Q5Y3 A0A1W4WFY5 A0A1B0FI70 A0A034WVB9 A0A0K8WG52 A0A0A1XD79 A0A0K8UF92 V5I904 A0A0K8UPM9 A0A182P2H4 B4NBQ3 D6WT89 A0A3L8D8F5 A0A084WQT7 A0A1Q3FUN9 B4KAU0 P36975 B3NEJ7 A0A1W4WG12 B4JHR0 A0A3B0KWI1 T1DGX4 A0A1L8DW05 B4GE78 A0A1J1J8C7 A0A195FF09 A0A2S2PNY8 A0A2H8TGS1 J9JTE9 A0A1L8DVZ5 Q17CJ4 A0A1Q3G430 A0A1B6D8D6 A0A1Y1JVQ4 B4MCB5 A0A1L8DVY9 A0A069DQM2 A0A1A9YSZ5 A0A1A9ZZ40 A0A023F5R8 A0A0T6AXP0 A0A1J1J428 A0A0Q9WT28 A0A0Q9WWQ8 A0A1B6L6U0 A0A0Q5UL58 P36975-3 A0A1B6F4I9 A0A2M4CTM3 A0A2M3Z6U4 A0A2M4AHI6 A0A1Q3G3Z9 I5AP65 A0A0P5E0U3 D3TKT6 A0A0Q9WJA9 A0A3B0KP36 A0A182L924 A0A182HKR3 E9H4F6 A0A1J1J425 A0A0A9Y7U7 A0A146M9E8 A0A310SJ16 A0A1B6CG89 T1E2Z7 A0A0M8ZVQ7 A0A182YGF5 A0A0L7R735 A0A182IJF9 A0A182N5D0 A0A182VR86 A0A154P3E2 A0A2J7QX39 A0A3Q0IS99 B4IVG1 A0A0R1E8J2 A0A1B6JVB9 A0A0P5EGM5 T1DC81
A0A1Y9IXP0 A0A1L8DVY8 A0A158NHW8 A0A1W4WS64 A0A1Y9IXY3 Q7PUS1 U5EIP5 E0VV38 A0A1Y9IY75 A0A1W7R879 A0A182RNH4 W8CEE5 A0A1W4WRW1 A0A1B6ENN8 A0A1I8Q5Y3 A0A1W4WFY5 A0A1B0FI70 A0A034WVB9 A0A0K8WG52 A0A0A1XD79 A0A0K8UF92 V5I904 A0A0K8UPM9 A0A182P2H4 B4NBQ3 D6WT89 A0A3L8D8F5 A0A084WQT7 A0A1Q3FUN9 B4KAU0 P36975 B3NEJ7 A0A1W4WG12 B4JHR0 A0A3B0KWI1 T1DGX4 A0A1L8DW05 B4GE78 A0A1J1J8C7 A0A195FF09 A0A2S2PNY8 A0A2H8TGS1 J9JTE9 A0A1L8DVZ5 Q17CJ4 A0A1Q3G430 A0A1B6D8D6 A0A1Y1JVQ4 B4MCB5 A0A1L8DVY9 A0A069DQM2 A0A1A9YSZ5 A0A1A9ZZ40 A0A023F5R8 A0A0T6AXP0 A0A1J1J428 A0A0Q9WT28 A0A0Q9WWQ8 A0A1B6L6U0 A0A0Q5UL58 P36975-3 A0A1B6F4I9 A0A2M4CTM3 A0A2M3Z6U4 A0A2M4AHI6 A0A1Q3G3Z9 I5AP65 A0A0P5E0U3 D3TKT6 A0A0Q9WJA9 A0A3B0KP36 A0A182L924 A0A182HKR3 E9H4F6 A0A1J1J425 A0A0A9Y7U7 A0A146M9E8 A0A310SJ16 A0A1B6CG89 T1E2Z7 A0A0M8ZVQ7 A0A182YGF5 A0A0L7R735 A0A182IJF9 A0A182N5D0 A0A182VR86 A0A154P3E2 A0A2J7QX39 A0A3Q0IS99 B4IVG1 A0A0R1E8J2 A0A1B6JVB9 A0A0P5EGM5 T1DC81
Pubmed
22118469
26354079
28756777
12364791
14747013
17210077
+ More
21347285 20566863 24495485 25348373 25830018 17994087 18362917 19820115 30249741 24438588 8226991 9272858 10731132 12537572 17510324 28004739 26334808 25474469 15632085 20353571 20966253 21292972 25401762 26823975 24330624 25244985 17550304
21347285 20566863 24495485 25348373 25830018 17994087 18362917 19820115 30249741 24438588 8226991 9272858 10731132 12537572 17510324 28004739 26334808 25474469 15632085 20353571 20966253 21292972 25401762 26823975 24330624 25244985 17550304
EMBL
AGBW02009942
OWR49529.1
KQ459604
KPI92339.1
KZ150014
PZC75011.1
+ More
NWSH01000170 PCG78791.1 AAAB01008987 EGK97596.1 GFDF01003492 JAV10592.1 ADTU01016089 ADTU01016090 ADTU01016091 ADTU01016092 ADTU01016093 EAA01106.5 GANO01002549 JAB57322.1 DS235802 EEB17244.1 GEHC01000322 JAV47323.1 GAMC01000336 JAC06220.1 GECZ01030233 JAS39536.1 CCAG010001331 GAKP01000676 GAKP01000675 JAC58277.1 GDHF01012096 GDHF01002173 JAI40218.1 JAI50141.1 GBXI01005377 JAD08915.1 GDHF01026972 JAI25342.1 GALX01003780 JAB64686.1 GDHF01023773 JAI28541.1 CH964232 EDW81217.1 KQ971354 EFA07177.2 QOIP01000012 RLU16208.1 ATLV01025728 ATLV01025729 ATLV01025730 KE525398 KFB52581.1 GFDL01003873 JAV31172.1 CH933806 EDW13613.1 L22021 U81153 U81147 U81148 U81149 U81150 U81151 U81152 AE014296 BT023789 BT126217 CH954178 EDV52832.1 CH916369 EDV93899.1 OUUW01000014 SPP88378.1 GAMD01002596 JAA98994.1 GFDF01003485 JAV10599.1 CH479182 EDW33913.1 CVRI01000070 CRL07161.1 KQ981636 KYN38807.1 GGMR01018540 MBY31159.1 GFXV01001117 MBW12922.1 ABLF02029848 ABLF02029852 GFDF01003486 JAV10598.1 CH477308 EAT44027.1 GFDL01000501 JAV34544.1 GEDC01015344 GEDC01010772 GEDC01003414 JAS21954.1 JAS26526.1 JAS33884.1 GEZM01099278 JAV53402.1 CH940657 EDW63197.2 GFDF01003484 JAV10600.1 GBGD01002898 JAC85991.1 GBBI01002192 JAC16520.1 LJIG01022610 KRT79645.1 CRL07159.1 KRF99406.1 KRG00502.1 GEBQ01020607 JAT19370.1 KQS44466.1 GECZ01028264 GECZ01025351 GECZ01024649 GECZ01018083 JAS41505.1 JAS44418.1 JAS45120.1 JAS51686.1 GGFL01004509 MBW68687.1 GGFM01003498 MBW24249.1 GGFK01006919 MBW40240.1 GFDL01000513 JAV34532.1 CM000070 EIM52750.2 GDIP01148710 JAJ74692.1 EZ422023 ADD18446.1 KRF80955.1 SPP88379.1 APCN01000860 GL732591 EFX73410.1 CRL07160.1 GBHO01018014 GBHO01002976 GBHO01002974 GBRD01011856 GDHC01002985 JAG25590.1 JAG40628.1 JAG40630.1 JAG53968.1 JAQ15644.1 GDHC01002238 JAQ16391.1 KQ764717 OAD54412.1 GEDC01030688 GEDC01024834 JAS06610.1 JAS12464.1 GALA01000840 JAA94012.1 KQ435848 KOX70996.1 KQ414643 KOC66658.1 KQ434809 KZC06367.1 NEVH01009395 PNF33157.1 CH892681 EDX00326.2 KRK05547.1 GECU01004853 JAT02854.1 GDIP01148709 GDIQ01254763 JAJ74693.1 JAJ96961.1 GAKT01000117 JAA92945.1
NWSH01000170 PCG78791.1 AAAB01008987 EGK97596.1 GFDF01003492 JAV10592.1 ADTU01016089 ADTU01016090 ADTU01016091 ADTU01016092 ADTU01016093 EAA01106.5 GANO01002549 JAB57322.1 DS235802 EEB17244.1 GEHC01000322 JAV47323.1 GAMC01000336 JAC06220.1 GECZ01030233 JAS39536.1 CCAG010001331 GAKP01000676 GAKP01000675 JAC58277.1 GDHF01012096 GDHF01002173 JAI40218.1 JAI50141.1 GBXI01005377 JAD08915.1 GDHF01026972 JAI25342.1 GALX01003780 JAB64686.1 GDHF01023773 JAI28541.1 CH964232 EDW81217.1 KQ971354 EFA07177.2 QOIP01000012 RLU16208.1 ATLV01025728 ATLV01025729 ATLV01025730 KE525398 KFB52581.1 GFDL01003873 JAV31172.1 CH933806 EDW13613.1 L22021 U81153 U81147 U81148 U81149 U81150 U81151 U81152 AE014296 BT023789 BT126217 CH954178 EDV52832.1 CH916369 EDV93899.1 OUUW01000014 SPP88378.1 GAMD01002596 JAA98994.1 GFDF01003485 JAV10599.1 CH479182 EDW33913.1 CVRI01000070 CRL07161.1 KQ981636 KYN38807.1 GGMR01018540 MBY31159.1 GFXV01001117 MBW12922.1 ABLF02029848 ABLF02029852 GFDF01003486 JAV10598.1 CH477308 EAT44027.1 GFDL01000501 JAV34544.1 GEDC01015344 GEDC01010772 GEDC01003414 JAS21954.1 JAS26526.1 JAS33884.1 GEZM01099278 JAV53402.1 CH940657 EDW63197.2 GFDF01003484 JAV10600.1 GBGD01002898 JAC85991.1 GBBI01002192 JAC16520.1 LJIG01022610 KRT79645.1 CRL07159.1 KRF99406.1 KRG00502.1 GEBQ01020607 JAT19370.1 KQS44466.1 GECZ01028264 GECZ01025351 GECZ01024649 GECZ01018083 JAS41505.1 JAS44418.1 JAS45120.1 JAS51686.1 GGFL01004509 MBW68687.1 GGFM01003498 MBW24249.1 GGFK01006919 MBW40240.1 GFDL01000513 JAV34532.1 CM000070 EIM52750.2 GDIP01148710 JAJ74692.1 EZ422023 ADD18446.1 KRF80955.1 SPP88379.1 APCN01000860 GL732591 EFX73410.1 CRL07160.1 GBHO01018014 GBHO01002976 GBHO01002974 GBRD01011856 GDHC01002985 JAG25590.1 JAG40628.1 JAG40630.1 JAG53968.1 JAQ15644.1 GDHC01002238 JAQ16391.1 KQ764717 OAD54412.1 GEDC01030688 GEDC01024834 JAS06610.1 JAS12464.1 GALA01000840 JAA94012.1 KQ435848 KOX70996.1 KQ414643 KOC66658.1 KQ434809 KZC06367.1 NEVH01009395 PNF33157.1 CH892681 EDX00326.2 KRK05547.1 GECU01004853 JAT02854.1 GDIP01148709 GDIQ01254763 JAJ74693.1 JAJ96961.1 GAKT01000117 JAA92945.1
Proteomes
UP000007151
UP000053268
UP000218220
UP000076407
UP000007062
UP000005205
+ More
UP000192223 UP000009046 UP000075900 UP000095300 UP000092444 UP000075885 UP000007798 UP000007266 UP000279307 UP000030765 UP000009192 UP000000803 UP000008711 UP000001070 UP000268350 UP000008744 UP000183832 UP000078541 UP000007819 UP000008820 UP000008792 UP000092443 UP000092445 UP000001819 UP000075882 UP000075840 UP000000305 UP000053105 UP000076408 UP000053825 UP000075880 UP000075884 UP000075920 UP000076502 UP000235965 UP000079169 UP000002282
UP000192223 UP000009046 UP000075900 UP000095300 UP000092444 UP000075885 UP000007798 UP000007266 UP000279307 UP000030765 UP000009192 UP000000803 UP000008711 UP000001070 UP000268350 UP000008744 UP000183832 UP000078541 UP000007819 UP000008820 UP000008792 UP000092443 UP000092445 UP000001819 UP000075882 UP000075840 UP000000305 UP000053105 UP000076408 UP000053825 UP000075880 UP000075884 UP000075920 UP000076502 UP000235965 UP000079169 UP000002282
PRIDE
Pfam
PF00835 SNAP-25
ProteinModelPortal
A0A212F742
A0A194PHY8
A0A2W1BL53
A0A2A4K3F5
A0A182X9T1
F5HMS4
+ More
A0A1Y9IXP0 A0A1L8DVY8 A0A158NHW8 A0A1W4WS64 A0A1Y9IXY3 Q7PUS1 U5EIP5 E0VV38 A0A1Y9IY75 A0A1W7R879 A0A182RNH4 W8CEE5 A0A1W4WRW1 A0A1B6ENN8 A0A1I8Q5Y3 A0A1W4WFY5 A0A1B0FI70 A0A034WVB9 A0A0K8WG52 A0A0A1XD79 A0A0K8UF92 V5I904 A0A0K8UPM9 A0A182P2H4 B4NBQ3 D6WT89 A0A3L8D8F5 A0A084WQT7 A0A1Q3FUN9 B4KAU0 P36975 B3NEJ7 A0A1W4WG12 B4JHR0 A0A3B0KWI1 T1DGX4 A0A1L8DW05 B4GE78 A0A1J1J8C7 A0A195FF09 A0A2S2PNY8 A0A2H8TGS1 J9JTE9 A0A1L8DVZ5 Q17CJ4 A0A1Q3G430 A0A1B6D8D6 A0A1Y1JVQ4 B4MCB5 A0A1L8DVY9 A0A069DQM2 A0A1A9YSZ5 A0A1A9ZZ40 A0A023F5R8 A0A0T6AXP0 A0A1J1J428 A0A0Q9WT28 A0A0Q9WWQ8 A0A1B6L6U0 A0A0Q5UL58 P36975-3 A0A1B6F4I9 A0A2M4CTM3 A0A2M3Z6U4 A0A2M4AHI6 A0A1Q3G3Z9 I5AP65 A0A0P5E0U3 D3TKT6 A0A0Q9WJA9 A0A3B0KP36 A0A182L924 A0A182HKR3 E9H4F6 A0A1J1J425 A0A0A9Y7U7 A0A146M9E8 A0A310SJ16 A0A1B6CG89 T1E2Z7 A0A0M8ZVQ7 A0A182YGF5 A0A0L7R735 A0A182IJF9 A0A182N5D0 A0A182VR86 A0A154P3E2 A0A2J7QX39 A0A3Q0IS99 B4IVG1 A0A0R1E8J2 A0A1B6JVB9 A0A0P5EGM5 T1DC81
A0A1Y9IXP0 A0A1L8DVY8 A0A158NHW8 A0A1W4WS64 A0A1Y9IXY3 Q7PUS1 U5EIP5 E0VV38 A0A1Y9IY75 A0A1W7R879 A0A182RNH4 W8CEE5 A0A1W4WRW1 A0A1B6ENN8 A0A1I8Q5Y3 A0A1W4WFY5 A0A1B0FI70 A0A034WVB9 A0A0K8WG52 A0A0A1XD79 A0A0K8UF92 V5I904 A0A0K8UPM9 A0A182P2H4 B4NBQ3 D6WT89 A0A3L8D8F5 A0A084WQT7 A0A1Q3FUN9 B4KAU0 P36975 B3NEJ7 A0A1W4WG12 B4JHR0 A0A3B0KWI1 T1DGX4 A0A1L8DW05 B4GE78 A0A1J1J8C7 A0A195FF09 A0A2S2PNY8 A0A2H8TGS1 J9JTE9 A0A1L8DVZ5 Q17CJ4 A0A1Q3G430 A0A1B6D8D6 A0A1Y1JVQ4 B4MCB5 A0A1L8DVY9 A0A069DQM2 A0A1A9YSZ5 A0A1A9ZZ40 A0A023F5R8 A0A0T6AXP0 A0A1J1J428 A0A0Q9WT28 A0A0Q9WWQ8 A0A1B6L6U0 A0A0Q5UL58 P36975-3 A0A1B6F4I9 A0A2M4CTM3 A0A2M3Z6U4 A0A2M4AHI6 A0A1Q3G3Z9 I5AP65 A0A0P5E0U3 D3TKT6 A0A0Q9WJA9 A0A3B0KP36 A0A182L924 A0A182HKR3 E9H4F6 A0A1J1J425 A0A0A9Y7U7 A0A146M9E8 A0A310SJ16 A0A1B6CG89 T1E2Z7 A0A0M8ZVQ7 A0A182YGF5 A0A0L7R735 A0A182IJF9 A0A182N5D0 A0A182VR86 A0A154P3E2 A0A2J7QX39 A0A3Q0IS99 B4IVG1 A0A0R1E8J2 A0A1B6JVB9 A0A0P5EGM5 T1DC81
PDB
6MDP
E-value=1.83848e-52,
Score=517
Ontologies
KEGG
GO
GO:0017075
GO:0005249
GO:0031201
GO:0006906
GO:0019905
GO:0098793
GO:0005886
GO:0016082
GO:0005484
GO:0031629
GO:0006887
GO:0006893
GO:0016192
GO:0005737
GO:0016081
GO:0000149
GO:0007269
GO:0007274
GO:0048172
GO:0048489
GO:0043195
GO:0030054
GO:0016020
GO:0005515
GO:0030001
GO:0046873
GO:0000287
GO:0043874
GO:0003677
GO:0008408
GO:0005112
GO:0015930
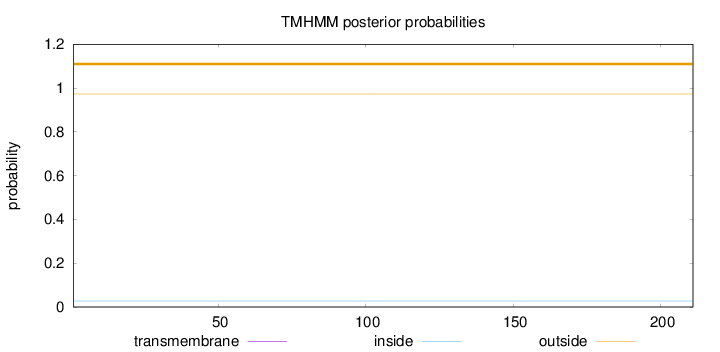
Topology
Subcellular location
Complexed with macromolecular elements of the nerve terminal. With evidence from 1 publications.
Length:
211
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02713
outside
1 - 211
Population Genetic Test Statistics
Pi
279.183269
Theta
179.627345
Tajima's D
1.335821
CLR
0.13381
CSRT
0.746162691865407
Interpretation
Uncertain
