Gene
KWMTBOMO15219
Pre Gene Modal
BGIBMGA004955
Annotation
PREDICTED:_zinc_transporter_ZIP1-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.902
Sequence
CDS
ATGGCCGTCGAAAACGTTTTAACCGCTAAAATAGTAACAATGGTATCCTTATTCGGGATATCCATGTTCGTGGGATGTTTGCCGTTGTTGATATCGAAGAAGTTCAACTGGTTCACGAAATCCCGCGATCCAAACCTGCGGTCTTCGAATCAGCTGGTGATGGGACTGTTGGCGTTTGGTGGCGGAGTACTTTTCGCTACAACGTTCATGCATCTCCTGCCTGAGGTGGACGAAAACATTGAATTGTTGCAAGAGACAGGAGAGATCCCCGATGTGCCAGTGTTCCTTGCCGGTTTGATAATGTGCGGAGGATTCTTCATGATGTACTTAGTCGAAGAATTAGTTCACTTGTACATACACGCCAGGGAGCACAAACCATCAGAGAAGAACGACGCCAAACCGTTCAGCCTAAGGAAGAGCATCTCGGGGAACGATCCAGAAATAAACCAAGAGACCAAACAAATTGAGAATGAAACCAATAGCGGTCATAGTCATTTAGTTCTCGATTCATCCCAAGAAGATTCTGTGGTGTCAGCCCTGAGAGGCTTGCTGATAGTCCTGGCGCTATCCATTCACGAACTGTTCGAAGGATTGGCAGTCGGCCTAGAGTCTTCAGTTGCCAATGTCTGGTACATGTTCGGAGCGGTGTCAGCCCACAAATACATCATTGCTTTCTGCATCGGAGTTGAACTGTTAGCGGCTGGCACGAGAAAATGGCTGTCTCTCGTCTACATCTTTACGTTTTCGTTCATGTCAGCCCTGGGTATCGGCATTGGGATCTTACTAGTAGGAGGGTCAGGGGCCGCGGCAGCTGGACTATATTCAGTGGTGTTACAGGGCATTGCCTGCGGTACTCTGGTCTACGTGGTGTTCTTTGAGGTGTGGAAGCGGAGCAGCGGTCTCCTGCAGTTCGCGTGCGCTCTCCTCGGGTTTGCGGTCATGGTTTCTCTCGAGGTTACCGTTGAATTCAGTGAGTAA
Protein
MAVENVLTAKIVTMVSLFGISMFVGCLPLLISKKFNWFTKSRDPNLRSSNQLVMGLLAFGGGVLFATTFMHLLPEVDENIELLQETGEIPDVPVFLAGLIMCGGFFMMYLVEELVHLYIHAREHKPSEKNDAKPFSLRKSISGNDPEINQETKQIENETNSGHSHLVLDSSQEDSVVSALRGLLIVLALSIHELFEGLAVGLESSVANVWYMFGAVSAHKYIIAFCIGVELLAAGTRKWLSLVYIFTFSFMSALGIGIGILLVGGSGAAAAGLYSVVLQGIACGTLVYVVFFEVWKRSSGLLQFACALLGFAVMVSLEVTVEFSE
Summary
Uniprot
A0A2W1BQK0
A0A2A4K3G4
A0A0L7L4V3
A0A194RKU1
A0A1E1WK27
A0A3S2LZ69
+ More
A0A212ENZ5 A0A2H1VSV6 A0A2W1BIW7 A0A194RJZ6 A0A1E1VXG4 A0A212F716 A0A0T6B976 A0A0L7QQF8 E2AVB2 A0A1Y1KHB2 A0A1B0D4L4 A0A154PFS4 A0A026WT16 A0A158NI77 A0A3L8DFW7 A0A195AWG9 K7INB1 A0A151WR48 E2C7U4 A0A3S2TJ53 A0A336LKY4 A0A2P8YQQ9 A0A310SKB6 A0A232FEZ9 B4J6J8 A0A139WLF5 U5EYZ1 A0A182GMX7 A0A195EGJ1 A0A195CDF2 N6TNY1 A0A023ERR6 Q17KP2 A0A336N2M7 A0A1S4EZC7 A0A195FFN2 F4WYB2 A0A1L8DEF2 B3NB30 A0A1L8DEK4 A0A1L8DEP6 B4Q4V3 A0A1J1HMH3 A0A1W4X9V4 A0A1W4XKA2 B4P105 A0A0C9R6C4 Q7JZR2 Q7K9B7 A0A067RIJ0 B4HQB1 A0A1S4J9S6 B3M2H8 A0A1L8DED9 A0A1L8DE33 A0A1L8DE30 B3P104 B0W7S1 A0A1I8P7F8 B4LK12 A0A0J7KKP8 A0A1Q3FD84 A0A1Q3FD12 B4PR16 N6TRJ9 T1GAW1 E9JB69 Q8SZF0 Q9GRW9 Q292Y9 B4GC82 A0A1W4VDS1 Q9VEX1 B3MIP2 A0A2A4IY73 A0A2J7QPU8 B4IBY1 B4QX54 B4K5S4 A0A088AA91 A0A3B0JQR8 A0A1W4WE97 A0A194PH19 B4MY51 B4KNI1 A0A3B0JII0 Q295U9 B4GFN4 H9J664 A0A0M4EDX2 A0A2J7QPT7 A0A2H1VCT4
A0A212ENZ5 A0A2H1VSV6 A0A2W1BIW7 A0A194RJZ6 A0A1E1VXG4 A0A212F716 A0A0T6B976 A0A0L7QQF8 E2AVB2 A0A1Y1KHB2 A0A1B0D4L4 A0A154PFS4 A0A026WT16 A0A158NI77 A0A3L8DFW7 A0A195AWG9 K7INB1 A0A151WR48 E2C7U4 A0A3S2TJ53 A0A336LKY4 A0A2P8YQQ9 A0A310SKB6 A0A232FEZ9 B4J6J8 A0A139WLF5 U5EYZ1 A0A182GMX7 A0A195EGJ1 A0A195CDF2 N6TNY1 A0A023ERR6 Q17KP2 A0A336N2M7 A0A1S4EZC7 A0A195FFN2 F4WYB2 A0A1L8DEF2 B3NB30 A0A1L8DEK4 A0A1L8DEP6 B4Q4V3 A0A1J1HMH3 A0A1W4X9V4 A0A1W4XKA2 B4P105 A0A0C9R6C4 Q7JZR2 Q7K9B7 A0A067RIJ0 B4HQB1 A0A1S4J9S6 B3M2H8 A0A1L8DED9 A0A1L8DE33 A0A1L8DE30 B3P104 B0W7S1 A0A1I8P7F8 B4LK12 A0A0J7KKP8 A0A1Q3FD84 A0A1Q3FD12 B4PR16 N6TRJ9 T1GAW1 E9JB69 Q8SZF0 Q9GRW9 Q292Y9 B4GC82 A0A1W4VDS1 Q9VEX1 B3MIP2 A0A2A4IY73 A0A2J7QPU8 B4IBY1 B4QX54 B4K5S4 A0A088AA91 A0A3B0JQR8 A0A1W4WE97 A0A194PH19 B4MY51 B4KNI1 A0A3B0JII0 Q295U9 B4GFN4 H9J664 A0A0M4EDX2 A0A2J7QPT7 A0A2H1VCT4
Pubmed
28756777
26227816
26354079
22118469
20798317
28004739
+ More
24508170 21347285 30249741 20075255 29403074 28648823 17994087 18362917 19820115 26483478 23537049 24945155 17510324 21719571 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 21282665 15632085 19121390
24508170 21347285 30249741 20075255 29403074 28648823 17994087 18362917 19820115 26483478 23537049 24945155 17510324 21719571 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 21282665 15632085 19121390
EMBL
KZ150014
PZC75010.1
NWSH01000170
PCG78801.1
JTDY01002941
KOB70445.1
+ More
KQ460045 KPJ18157.1 GDQN01003787 JAT87267.1 RSAL01000102 RVE47486.1 AGBW02013585 OWR43199.1 ODYU01004243 SOQ43910.1 PZC75012.1 KPJ18153.1 GDQN01011647 JAT79407.1 AGBW02009942 OWR49531.1 LJIG01009022 KRT83912.1 KQ414786 KOC60850.1 GL443059 EFN62666.1 GEZM01084004 GEZM01084003 JAV60859.1 AJVK01024657 KQ434896 KZC10726.1 KK107109 EZA59147.1 ADTU01016065 QOIP01000008 RLU19196.1 KQ976725 KYM76593.1 KQ982813 KYQ50277.1 GL453466 EFN75979.1 RVE47484.1 UFQT01000005 SSX17319.1 PYGN01000427 PSN46581.1 KQ765605 OAD53872.1 NNAY01000318 OXU29265.1 CH916367 EDW01999.1 KQ971321 KYB28661.1 GANO01000007 JAB59864.1 JXUM01075351 KQ562882 KXJ74939.1 KQ978957 KYN27266.1 KQ978023 KYM98098.1 APGK01057486 APGK01057487 KB741280 KB632033 ENN70945.1 ERL88183.1 GAPW01001720 JAC11878.1 CH477222 EAT47262.1 UFQT01003778 SSX35273.1 KQ981636 KYN38834.1 GL888439 EGI60869.1 GFDF01009263 JAV04821.1 CH954177 EDV59795.1 GFDF01009304 JAV04780.1 GFDF01009264 JAV04820.1 CM000361 CM002911 EDX05775.1 KMY91782.1 CVRI01000010 CRK88730.1 CM000157 EDW89079.2 GBYB01002301 JAG72068.1 AY071095 AE013599 AAL48717.1 AAM70819.2 AJ401614 CAC14873.1 KK852483 KDR22853.1 CH480816 EDW46650.1 CH902617 EDV43431.1 GFDF01009390 JAV04694.1 GFDF01009372 JAV04712.1 GFDF01009373 JAV04711.1 CH954181 EDV49123.1 DS231855 EDS38215.1 CH940648 EDW61666.1 LBMM01006206 KMQ90809.1 GFDL01009597 JAV25448.1 GFDL01009598 JAV25447.1 CM000160 EDW97338.1 ENN70946.1 CAQQ02197592 GL770938 EFZ09898.1 AY070921 AAL48543.1 AJ401615 CAC14874.1 CM000071 EAL24722.2 CH479181 EDW31400.1 AE014297 BT133201 AAF55295.1 AFA28442.1 CH902619 EDV38118.1 NWSH01005677 PCG64063.1 NEVH01012087 PNF30604.1 CH480827 EDW44889.1 CM000364 EDX12708.1 CH933806 EDW15136.1 OUUW01000008 SPP84517.1 KQ459604 KPI92343.1 CH963894 EDW77040.1 CH933808 EDW08940.2 OUUW01000001 SPP73026.1 CM000070 EAL28609.2 CH479182 EDW34419.1 BABH01019992 CP012524 ALC42177.1 PNF30595.1 ODYU01001850 SOQ38625.1
KQ460045 KPJ18157.1 GDQN01003787 JAT87267.1 RSAL01000102 RVE47486.1 AGBW02013585 OWR43199.1 ODYU01004243 SOQ43910.1 PZC75012.1 KPJ18153.1 GDQN01011647 JAT79407.1 AGBW02009942 OWR49531.1 LJIG01009022 KRT83912.1 KQ414786 KOC60850.1 GL443059 EFN62666.1 GEZM01084004 GEZM01084003 JAV60859.1 AJVK01024657 KQ434896 KZC10726.1 KK107109 EZA59147.1 ADTU01016065 QOIP01000008 RLU19196.1 KQ976725 KYM76593.1 KQ982813 KYQ50277.1 GL453466 EFN75979.1 RVE47484.1 UFQT01000005 SSX17319.1 PYGN01000427 PSN46581.1 KQ765605 OAD53872.1 NNAY01000318 OXU29265.1 CH916367 EDW01999.1 KQ971321 KYB28661.1 GANO01000007 JAB59864.1 JXUM01075351 KQ562882 KXJ74939.1 KQ978957 KYN27266.1 KQ978023 KYM98098.1 APGK01057486 APGK01057487 KB741280 KB632033 ENN70945.1 ERL88183.1 GAPW01001720 JAC11878.1 CH477222 EAT47262.1 UFQT01003778 SSX35273.1 KQ981636 KYN38834.1 GL888439 EGI60869.1 GFDF01009263 JAV04821.1 CH954177 EDV59795.1 GFDF01009304 JAV04780.1 GFDF01009264 JAV04820.1 CM000361 CM002911 EDX05775.1 KMY91782.1 CVRI01000010 CRK88730.1 CM000157 EDW89079.2 GBYB01002301 JAG72068.1 AY071095 AE013599 AAL48717.1 AAM70819.2 AJ401614 CAC14873.1 KK852483 KDR22853.1 CH480816 EDW46650.1 CH902617 EDV43431.1 GFDF01009390 JAV04694.1 GFDF01009372 JAV04712.1 GFDF01009373 JAV04711.1 CH954181 EDV49123.1 DS231855 EDS38215.1 CH940648 EDW61666.1 LBMM01006206 KMQ90809.1 GFDL01009597 JAV25448.1 GFDL01009598 JAV25447.1 CM000160 EDW97338.1 ENN70946.1 CAQQ02197592 GL770938 EFZ09898.1 AY070921 AAL48543.1 AJ401615 CAC14874.1 CM000071 EAL24722.2 CH479181 EDW31400.1 AE014297 BT133201 AAF55295.1 AFA28442.1 CH902619 EDV38118.1 NWSH01005677 PCG64063.1 NEVH01012087 PNF30604.1 CH480827 EDW44889.1 CM000364 EDX12708.1 CH933806 EDW15136.1 OUUW01000008 SPP84517.1 KQ459604 KPI92343.1 CH963894 EDW77040.1 CH933808 EDW08940.2 OUUW01000001 SPP73026.1 CM000070 EAL28609.2 CH479182 EDW34419.1 BABH01019992 CP012524 ALC42177.1 PNF30595.1 ODYU01001850 SOQ38625.1
Proteomes
UP000218220
UP000037510
UP000053240
UP000283053
UP000007151
UP000053825
+ More
UP000000311 UP000092462 UP000076502 UP000053097 UP000005205 UP000279307 UP000078540 UP000002358 UP000075809 UP000008237 UP000245037 UP000215335 UP000001070 UP000007266 UP000069940 UP000249989 UP000078492 UP000078542 UP000019118 UP000030742 UP000008820 UP000078541 UP000007755 UP000008711 UP000000304 UP000183832 UP000192223 UP000002282 UP000000803 UP000027135 UP000001292 UP000007801 UP000002320 UP000095300 UP000008792 UP000036403 UP000015102 UP000001819 UP000008744 UP000192221 UP000235965 UP000009192 UP000005203 UP000268350 UP000053268 UP000007798 UP000005204 UP000092553
UP000000311 UP000092462 UP000076502 UP000053097 UP000005205 UP000279307 UP000078540 UP000002358 UP000075809 UP000008237 UP000245037 UP000215335 UP000001070 UP000007266 UP000069940 UP000249989 UP000078492 UP000078542 UP000019118 UP000030742 UP000008820 UP000078541 UP000007755 UP000008711 UP000000304 UP000183832 UP000192223 UP000002282 UP000000803 UP000027135 UP000001292 UP000007801 UP000002320 UP000095300 UP000008792 UP000036403 UP000015102 UP000001819 UP000008744 UP000192221 UP000235965 UP000009192 UP000005203 UP000268350 UP000053268 UP000007798 UP000005204 UP000092553
Pfam
PF02535 Zip
Interpro
IPR003689
ZIP
ProteinModelPortal
A0A2W1BQK0
A0A2A4K3G4
A0A0L7L4V3
A0A194RKU1
A0A1E1WK27
A0A3S2LZ69
+ More
A0A212ENZ5 A0A2H1VSV6 A0A2W1BIW7 A0A194RJZ6 A0A1E1VXG4 A0A212F716 A0A0T6B976 A0A0L7QQF8 E2AVB2 A0A1Y1KHB2 A0A1B0D4L4 A0A154PFS4 A0A026WT16 A0A158NI77 A0A3L8DFW7 A0A195AWG9 K7INB1 A0A151WR48 E2C7U4 A0A3S2TJ53 A0A336LKY4 A0A2P8YQQ9 A0A310SKB6 A0A232FEZ9 B4J6J8 A0A139WLF5 U5EYZ1 A0A182GMX7 A0A195EGJ1 A0A195CDF2 N6TNY1 A0A023ERR6 Q17KP2 A0A336N2M7 A0A1S4EZC7 A0A195FFN2 F4WYB2 A0A1L8DEF2 B3NB30 A0A1L8DEK4 A0A1L8DEP6 B4Q4V3 A0A1J1HMH3 A0A1W4X9V4 A0A1W4XKA2 B4P105 A0A0C9R6C4 Q7JZR2 Q7K9B7 A0A067RIJ0 B4HQB1 A0A1S4J9S6 B3M2H8 A0A1L8DED9 A0A1L8DE33 A0A1L8DE30 B3P104 B0W7S1 A0A1I8P7F8 B4LK12 A0A0J7KKP8 A0A1Q3FD84 A0A1Q3FD12 B4PR16 N6TRJ9 T1GAW1 E9JB69 Q8SZF0 Q9GRW9 Q292Y9 B4GC82 A0A1W4VDS1 Q9VEX1 B3MIP2 A0A2A4IY73 A0A2J7QPU8 B4IBY1 B4QX54 B4K5S4 A0A088AA91 A0A3B0JQR8 A0A1W4WE97 A0A194PH19 B4MY51 B4KNI1 A0A3B0JII0 Q295U9 B4GFN4 H9J664 A0A0M4EDX2 A0A2J7QPT7 A0A2H1VCT4
A0A212ENZ5 A0A2H1VSV6 A0A2W1BIW7 A0A194RJZ6 A0A1E1VXG4 A0A212F716 A0A0T6B976 A0A0L7QQF8 E2AVB2 A0A1Y1KHB2 A0A1B0D4L4 A0A154PFS4 A0A026WT16 A0A158NI77 A0A3L8DFW7 A0A195AWG9 K7INB1 A0A151WR48 E2C7U4 A0A3S2TJ53 A0A336LKY4 A0A2P8YQQ9 A0A310SKB6 A0A232FEZ9 B4J6J8 A0A139WLF5 U5EYZ1 A0A182GMX7 A0A195EGJ1 A0A195CDF2 N6TNY1 A0A023ERR6 Q17KP2 A0A336N2M7 A0A1S4EZC7 A0A195FFN2 F4WYB2 A0A1L8DEF2 B3NB30 A0A1L8DEK4 A0A1L8DEP6 B4Q4V3 A0A1J1HMH3 A0A1W4X9V4 A0A1W4XKA2 B4P105 A0A0C9R6C4 Q7JZR2 Q7K9B7 A0A067RIJ0 B4HQB1 A0A1S4J9S6 B3M2H8 A0A1L8DED9 A0A1L8DE33 A0A1L8DE30 B3P104 B0W7S1 A0A1I8P7F8 B4LK12 A0A0J7KKP8 A0A1Q3FD84 A0A1Q3FD12 B4PR16 N6TRJ9 T1GAW1 E9JB69 Q8SZF0 Q9GRW9 Q292Y9 B4GC82 A0A1W4VDS1 Q9VEX1 B3MIP2 A0A2A4IY73 A0A2J7QPU8 B4IBY1 B4QX54 B4K5S4 A0A088AA91 A0A3B0JQR8 A0A1W4WE97 A0A194PH19 B4MY51 B4KNI1 A0A3B0JII0 Q295U9 B4GFN4 H9J664 A0A0M4EDX2 A0A2J7QPT7 A0A2H1VCT4
Ontologies
GO
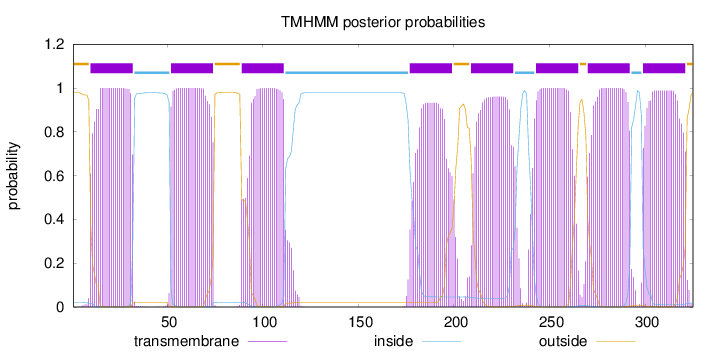
Topology
Length:
325
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
174.36992
Exp number, first 60 AAs:
30.80358
Total prob of N-in:
0.02122
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 51
TMhelix
52 - 74
outside
75 - 88
TMhelix
89 - 111
inside
112 - 176
TMhelix
177 - 199
outside
200 - 208
TMhelix
209 - 231
inside
232 - 242
TMhelix
243 - 265
outside
266 - 269
TMhelix
270 - 292
inside
293 - 298
TMhelix
299 - 321
outside
322 - 325
Population Genetic Test Statistics
Pi
254.09325
Theta
2.345011
Tajima's D
1.635024
CLR
0.273688
CSRT
0.852757362131893
Interpretation
Uncertain
